10 Haemosporida (Order): The “Malaria Parasites”
Susan L. Perkins and Spencer C. Galen
Phylum Myzozoa
Subphylum Apicomplexa
Order Haemosporida
Introduction
The term malaria refers specifically to a disease of humans that is caused by an infection of red blood cells (erythrocytes) and other cells by protozoan parasites in the genus Plasmodium and transmitted by mosquitoes in the genus Anopheles. The pathology of this disease results from 3 primary sources: 1) Episodic fevers that are caused by the cyclic rupturing of host erythrocytes; 2) anemia that follows from infection of host erythrocytes and their subsequent death; and 3) clogged capillaries from the combination of a loss of elasticity of infected erythrocytes as well as parasites triggering host erythrocytes to express surface proteins that make them likely to cling to one another. Nearly half a million people still die from malaria worldwide every year (Cibulskis et al., 2016; WHO, 2021), primarily in sub-Saharan Africa and other tropical regions. No vaccine is yet available on a global scale, but an RTS,S vaccine against the most pathogenic species (Plasmodium falciparum) has recently been approved and is being deployed in Ghana (WHO, 2023). Several drugs have been used as prophylaxis or treatment, but the parasites have evolved resistance to these compounds in many regions of the world. The human parasites are only a tiny fraction of the very diverse clade Haemosporida (sometimes called Haemospororida) which contains over 600 described species of these protozoan parasites occurring in many different species of reptiles, birds, and other mammals. All haemosporidians use a vertebrate host and a biting dipteran (fly) vector during different stages of their life cycles. Non-human haemosporidians are sometimes colloquially referred to as the malaria parasites, due to their close relationships and similar life cycle to the parasites that cause this disease in humans.
History of Knowledge of Malaria
Symptoms of malaria, particularly its regularly spaced fevers, have been described by writers in antiquity going back as far as 5,000 BCE in China and more than 3,000 years ago in India, Sumeria, and Egypt. The writings of ancient Greece describe characteristic symptoms of Plasmodium falciparum, P. vivax, and P. malariae and Alexander the Great is believed to have died from P. falciparum while attempting to travel to India in 323 BCE (Carter and Mendis, 2002). It is thought that both European colonists and the enslaved West Africans that they transported to the New World brought malaria parasites, and by the mid-19th century, malaria was a common and endemic disease throughout the tropical and temperate regions of North America and South America (Carter and Mendis, 2002).
Despite the ubiquity and severity of malaria, the root cause of this disease remained largely a mystery, with only the link between smelly, swampy regions and the resulting symptoms as a clue (the word malaria stems from the Italian words for bad air, namely, mal + aria). Eventually, in the late 1800s a series of scientists, most notably Charles Laveran, began to piece together that tiny specks in the blood of sick humans, later known to be the blood stages of the parasites, were associated with the characteristic fevers of the disease. How it could pass from one human to another was not known until 1897, when Ronald Ross (Figure 1), a British Army doctor, showed that there were cells that could be found in the saliva of Anopheles mosquitoes that had fed on birds that were somewhat similar to those that he observed in the blood of sick human patients. Ross was awarded the Nobel Prize in Physiology or Medicine in 1902 for this major piece of the discovery, but the other aspects of the malaria life cycle remained unknown for many decades to come, namely, where the sporozoites went between the time they were injected into the host by the mosquito and when they appeared in the bloodstream of the same host.
Malaria Today
Today, malaria is still one of the largest public health burdens in the world (Sachs and Malaney, 2002; WHO, 2021; 2023) with as many as a quarter of a billion new cases arising each year. The number of cases and particularly the number of deaths has fallen recently due to improved prevention measures such as insecticide-infused bed nets, better diagnostics, and improved treatments, however, there is still plenty of reason to be cautious. The drug artemisinin (and its derivatives), which is the first line of treatment in all malaria endemic-countries, is now at risk of lower efficacy as resistance has evolved in several endemic regions and insecticide resistance remains a looming and potential problem.
General Life Cycle
All members of the order Haemosporida follow the same generalized life cycle, which is obligately heteroxenous (multiple hosts), alternating between a vertebrate and a biting fly such as a mosquito, midge, louse fly, sand fly, or black fly (Figure 1). Take as an example the bite of an already infected insect (Figure 1, point 1). Several groups of flies use blood as a source of proteins and lipids with which to make their eggs. If a fly is infected with haemosporidians, when she feeds from the vertebrate, the stages of the parasite known as sporozoites will be injected into the vertebrate bloodstream along with her saliva. Sporozoites will travel through the bloodstream until they come to the liver or other target tissues, where they will invade the host cell and begin to asexually divide (Figure 1, point 2), often making thousands of daughter cells within just a few days. In some groups of haemosporidians, namely the genus Plasmodium, after this first round of tissue schizogony some of the parasites will leave the tissue stages and invade blood cells (typically red blood cells; Figure 1, point 3), where they transition into the asexual feeding stages known as trophozoites. During this point in the life cycle, the parasites digest the hemoglobin present in the hosts’ red blood cells, eventually forming a crystalline compound known as hemozoin. These cells will then undergo additional asexual division and burst from the host cell, going on to infect new blood cells. However, the majority of the genera in the order Haemosporida do not asexually replicate in the host’s blood cells, but instead continue their cycle in other tissues such as the epithelium of the lungs or spleen. All members of the order will at some point show the transmission stages, known as gametocytes, in the host blood cells (Figure 1, point 4). These cells exist as different sexes; females are known as macrogametocytes and males are known as microgametocytes. When another biting fly comes to feed on this host, the gametocytes will be taken up as part of her blood meal (Figure 1, point 5). Within the midgut of the fly, the gametocytes undergo a process called exflagellation, as the blood cells themselves begin to be digested. The microgametocytes split into several smaller microgametes, each of which can fertilize an exposed macrogamete (Figure 1, point 6). Once fused, the parasite exists as a motile stage called an ookinete (Figure 1, point 7). These cells push through the cells of the insect’s gut and encyst on the outer edge, in a structure called the oocyst (Figure 1, point 8). Thousands of sporozoites emerge from each oocyst and migrate to the insect’s salivary glands, where they wait until her next blood meal to infect a new vertebrate host (Figure 1, point 9).
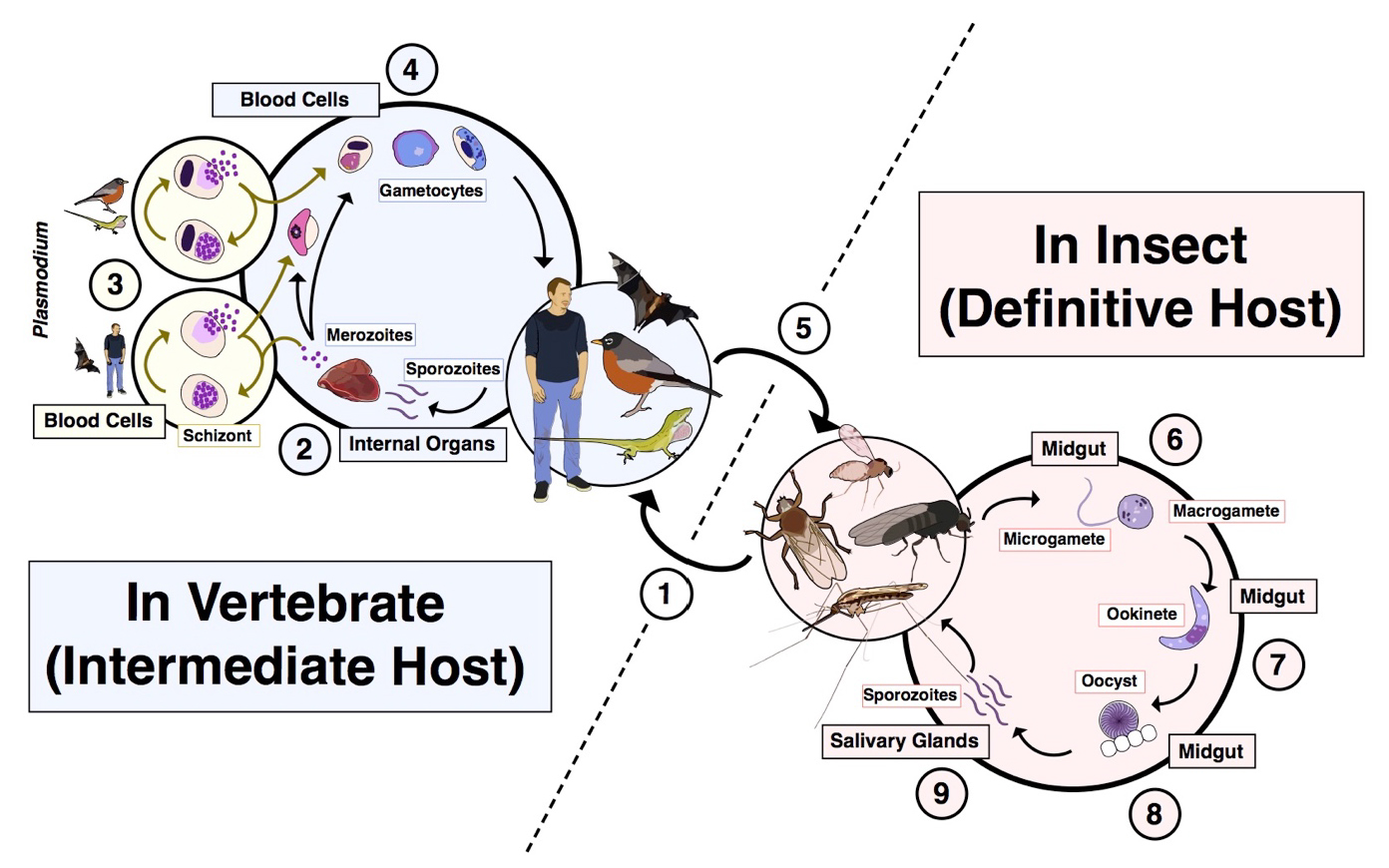
Figure 1. Generalized life cycle for haemosporidian (malaria) parasites. 1) An infected dipteran takes a blood meal from a vertebrate and sporozoite stages are injected into the bloodstream along with its saliva. 2) The sporozoites travel through the blood until they reach specific cells in internal organs where they will undergo rounds of asexual division. In human malaria parasites and many others of mammals, this occurs in cells of the liver. 3) For Plasmodium parasites, there are additional rounds of asexual division in blood cells as well. This can occur in both anucleated red blood cells of mammals (bottom) and in nucleated blood cells of birds and squamates (top). 4) Eventually, a developmental switch triggers the development of gametocytes, or the transmission stages. These stages are either of future male function (microgametocytes) or female function (macrogametocytes). These stages are highly variable across the diversity of haemosporidians. The illustration depicts gametocytes of Plasmodium falciparum (lower left), a lizard Plasmodium (second from left), a Leucocytozoon (third from left), and a Haemoproteus (fourth from left). 5) If a dipteran vector feeds from an infected vertebrate host, she will pick up gametocytes as part of that blood meal. 6). Once inside the fly’s midgut, the gametes undergo exflagellation, forming the macrogamete (female) and multiple microgametes (male). 7) After fertilization, the zygote transforms into a motile stage known as the ookinete, which penetrates the wall of the fly’s midgut. 8) A cyst, known as the oocyst forms and meiosis occurs at this point. 9) Sporozoites rupture from the oocyst and travel to the fly’s head, coming to reside in the salivary glands to await the next blood meal.
(Source: S. C. Galen, 2019. License: CC BY-NC-SA 4.0.)
Because the sexual component of the malaria parasite’s life cycle occurs within the insect, technically it is the insect that should be referred to as the definitive host, with the human or other vertebrate host referred to as the intermediate host. The dipteran insect is, however, often referred to as the vector of the malaria parasite as it does transmit the parasite between humans or other vertebrate hosts. The complete life cycle has been worked out in detail for only a few of the species of haemosporidians that infect non-human hosts, with most assumed to follow the same stages as their close relatives in the same genus. Although the use of different insect host groups (Table 1) is known for some of these parasites, the vast majority of haemosporidians are transmitted by unknown species of flies.
Table 1. Major groups of haemosporidians, with vertebrate and dipteran hosts used.

Diversity of and Relationships within the Family Haemosporida
The species of Plasmodium that commonly infect humans are the best known and most intensively studied haemosporidians, though they are of very minor importance in terms of the overall diversity of these parasites. Birds or their dinosaur precursors are most likely the original vertebrate hosts of the malaria parasites and still harbor the greatest diversity of the haemosporidians, both in terms of the number of described species that use birds as hosts, but also their geographic range (Valkiūnas, 2004). Numerous other vertebrate groups are hosts to species of haemosporidians, including monkeys, bats, lizards, and turtles. Haemosporidians are typically not capable of infecting hosts outside of their major host group, that is, bird malaria parasites cannot infect humans and vice versa. However, within their major host groups some malaria parasites (especially the avian malaria parasites) can infect many different species. For example, the cosmopolitan avian malaria parasite P. relictum has been recorded to infect over 300 different species of birds throughout the world (Valkiūnas et al., 2018).
Classification
The classification of the malaria parasites has been extremely fluid throughout history. For the bulk of this time, taxonomic groups including genera and families were primarily structured around 2 primary characteristics of the parasite species: 1) If the parasite reproduced asexually in the host blood (known as erythrocytic schizogony) and 2) whether or not hemozoin pigment was visible in the blood stages of the parasite. These traits were considered to be the most important in the separation of the parasites into genera and served as the basis for the creation of 4 families within the order (Levine, 1988).
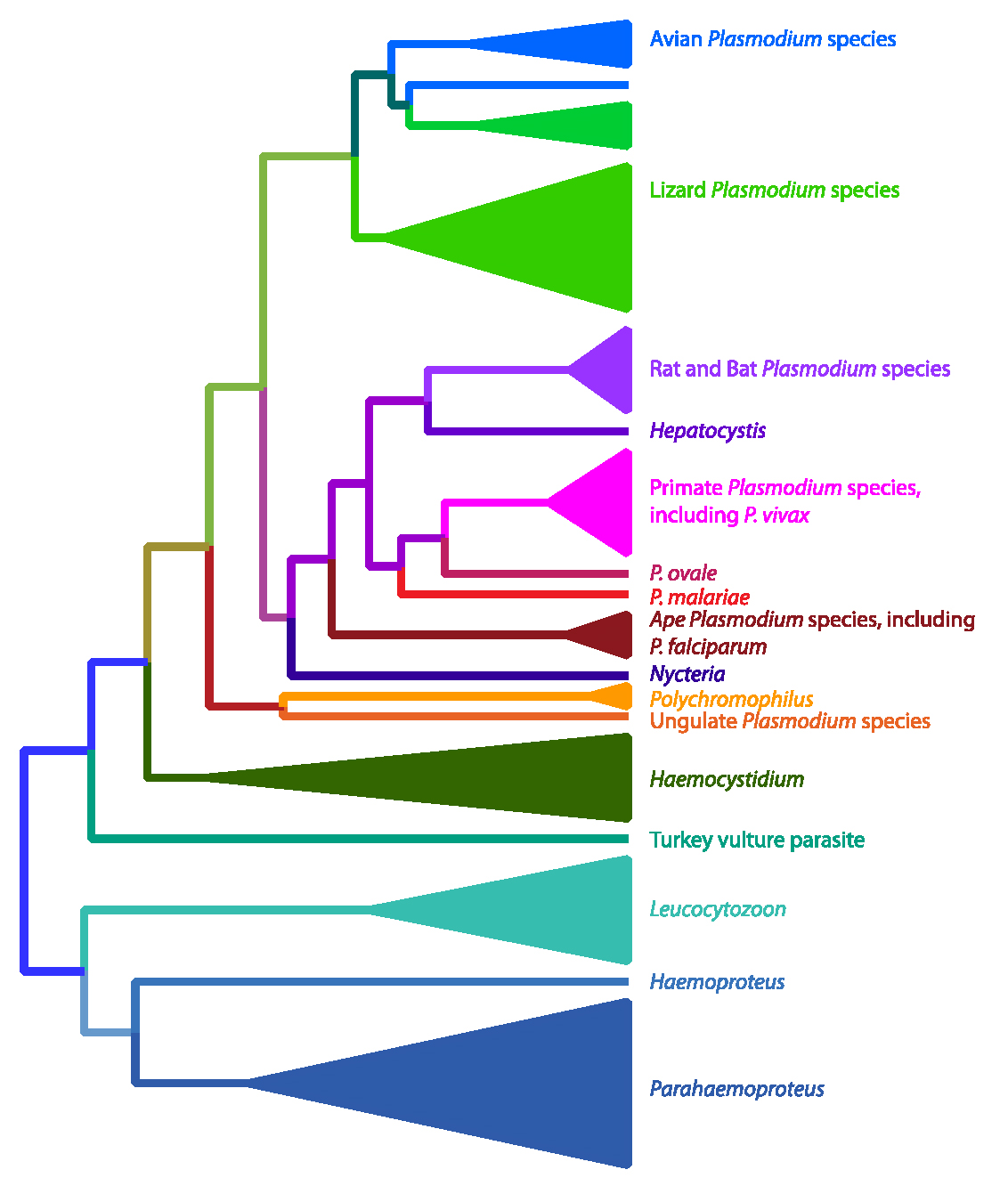
Figure 2. Phylogenetic relationships of the haemosporidian (malaria) parasites. An analysis of more than 20 genes (Galen et al., 2018) resulted in this hypothesis of the evolutionary relationships of various clades of haemosporidians. This tree supports that birds were the original hosts of these parasites with either a single introduction into mammalian hosts and a subsequent reinfection of birds and lizards by Plasmodium parasites, or that there were 2 invasions into mammals. This tree would make the genus Plasmodium polyphyletic as not all members share a common ancestor, however this would mean that significant taxonomic changes need to occur.
(Source: Galen et al., 2018. License: CC BY-NC-SA 4.0.)
The advent of using DNA sequences as characters with which to understand the evolutionary history and relationships among organisms drastically changed the hypotheses of the relationships within Haemosporida. The first molecular systematic study of these parasites used the 18S ribosomal small-subunit gene and a small set of taxa including the important human parasites, Plasmodium falciparum and P. vivax, as well as parasites found in rodents and 2 species from birds (Waters et al., 1991). The resulting topology showed a close relationship between P. falciparum and P. gallinaceum, a parasite that infects chickens, and the authors naturally concluded that humans had acquired the virulent P. falciparum following a host switch after chickens were domesticated. However, subsequent studies, particularly those using other genes, did not support the human/chicken connection (Ayala et al., 1999; Perkins and Schall, 2002) and instead showed that the human- and bird-infecting Plasmodium lineages are distantly related. These later studies also established that the avian parasites in the genus Leucocytozoon were likely an early-diverging lineage (Perkins and Schall, 2002). Recently, multiple genes from a large number of different haemosporidian parasites were sequenced and used to create the most comprehensive phylogeny to date (Figure 2) (Galen et al., 2018). These results highlighted the complex evolution of the Haemosporida and show that the original characters used to define clades have likely evolved more than once. This updated phylogeny also showed that the taxonomy of this group of parasites needed to be revised. For instance, most recent analyses have recovered parasites that have been classified as the distinct genus Hepatocystis as closely related to the human-infecting Plasmodium species (Perkins and Schall, 2002; Galen et al., 2018). Conversely, many parasites classified as Plasmodium because they show schizogony in blood cells and clear hemozoin pigment, have been shown not to be part of a monophyletic group that contains the other Plasmodium species, including the type species of the genus.
Malaria Parasites of Birds
Malaria parasites are practically ubiquitous in birds with a cosmpolitan distribution. Several hundred species have been described from the genera Plasmodium, Haemoproteus, Parahaemoproteus, and Leucocytozoon (Table 1; Figure 2), making the bird-infecting malaria parasites the most species rich group within the Haemosporida.
Avian malaria has been instrumental in studies of the disease ever since it was first discovered in the late 1800s. Ronald Ross (Figure 3), who won the Nobel Prize for his discovery that mosquitoes were responsible for transmitting the parasites from person to person, first did experiments on birds infected with Plasmodium parasites (Rivero and Gandon, 2018). Bird systems were also what allowed the discovery that the parasite first completes 1 or more exoerythrocytic stages before it begins to infect the blood cells of the host (Huff and Coulston, 1946). Also, experiments using avian malaria were useful for understanding immunity to Plasmodium infection by testing the efficacy of early anti-malarial drugs (Tonkin and Hawking, 1947; Rivero and Gandon, 2018). The method of inoculating naive hosts with sporozoite stages that had been rendered inactive, one that is being tested in humans now (2019), was first developed in an avian malaria system (Rivero and Gandon, 2018). A large number of researchers continue to use avian malaria as a model system for studying parasite-host interactions and diversification. The attractiveness of avian malaria as a system lies in the fact that it is relatively easy and cost-effective to sample large numbers of birds from a variety of species in a given habitat via mist-netting and drawing a small blood sample. Haemosporidian-specific primers are available that allow the samples to be rapidly screened for the presence of parasites and identified to lineages by sequencing. Comprehensive and publicly accessible databases can then be assembled (Bensch et al., 2009) so that comparative studies of host use and diversification are possible. Through this type of molecular work on avian systems, over 3,000 different lineages of malaria parasites have been reported from all over the world, with some authors estimating that the number of species of avian malaria parasites may be as high as 10,000 (Bensch et al., 2004). One pattern that has emerged from this work is that avian malaria parasites can exhibit host generalism (with a broad host range), infecting large numbers of distantly related species of birds, or host specialization (with a narrow host range), infecting a small number of closely related host species (Martínez-de la Puente et al., 2011; Svensson-Coelho et al., 2014; Ellis et al., 2015). The reasons for the higher abundance, diversity, and variation in host infection patterns exhibited by the avian malaria parasites relative to other malaria parasites are poorly understood and have led to an increased interest in avian haemosporidian research in recent years (Bensch et al., 2009).

Figure 3. Ronald Ross. Sir Ronald Ross (1857–1932) was a British medical doctor whose work in India on both avian and human malaria parasites resulted in the discovery that mosquitoes transmit infective stages between vertebrates. He won the Nobel Prize in Medicine in 1902.
(Photo source: United States National Library of Medicine Digital Collections, https://collections.nlm.nih.gov/catalog/nlm:nlmuid-101427700-img. Public domain.)
Although important in early laboratory studies of malaria, the popularity of using birds as a model system waned substantially when the rodent malaria parasites were successfully cycled in laboratory mice. However, because the rodent malaria system involves just a small set of closely related parasite species that are used to infect an unnatural host species, there has been a recent resurgence in using birds as experimental systems with which to study the biology of malaria parasites (Rivero and Gandon, 2018). These studies have been accelerated by the ability to sequence the first genomes of avian malaria parasites (Bensch et al., 2016; Lutz et al., 2016a; Böhme et al., 2018) as well as transcriptome studies that can be done quite easily in bird hosts (Videvall et al., 2015; Weinberg et al., 2018).
Examples of some of the questions that are easily addressed using avian malaria parasites include studies on parasite virulence in relation to parasitemia in the host (Palinauskas et al., 2018) and costs to the reproduction and survival of parasites in the mosquito vector (Pigeault et al., 2015; Yan et al., 2018; Vézilier et al., 2012). One of the most exciting recent discoveries involving avian malaria parasites was that infected birds showed a marked shortening in their telomeres, the ends of chromosomes, which are thought to be related to life span in vertebrates in general (Asghar et al., 2016; Remot et al., 2022). This discovery prompted similar examination of telomere length in malaria-infected humans and showed that cell-death is induced by Plasmodium infection in our species as well (Asghar et al., 2017).
Avian malaria parasites have also been shown to have negative impacts on naïve host populations in at least one tragic case where the parasite was accidentally introduced to a region. In the early 1800s, Culex mosquitoes were accidentally introduced to the Hawaiian Islands and a few decades later, Plasmodium relictum was also brought there. An endemic transmission cycle was established, which quickly spilled into the native Hawaiian avifauna and likely contributed to their extinctions of some species (van Riper et al., 1986; Atkinson and Samuel, 2010; Samuel, et al., 2011).
Malaria Parasites of Squamate Reptiles (Class Reptilia: Order Squamata)
As with birds, the malaria parasites of reptiles are also geographically widespread (occurring on every continent except for Antarctica) and diverse, with over 100 described species (Telford, 2008). They infect a large number of squamates as hosts including over 10 different families of lizards and 3 species in snakes.
In lizards, the pathology of haemosporidians has only been well studied in 2 different systems, with varying results. In western fence lizards (Sceloporus occidentalis) that are infected with Plasmodium mexicanum, serious fitness consequences from infection were observed (Schall, 1990). Male lizards with these parasites were less likely to be able to defend a territory and infected female lizards laid fewer eggs per clutch. However, in another system, the Saban anole and its Plasmodium parasites in the Caribbean, these results were not found—infected and uninfected lizards showed similar reproductive success and survival (Schall and Staats, 2002). The malaria parasites of lizards have been used as a model system to study a variety of host-parasite relationships, including the role of these parasites on sexual selection (Schall, 1983; Schall and Staats, 1997), the evolution of sex ratios for optimal transmission (Schall, 2000; 2009; Osgood and Schall, 2004; Neal and Schall, 2010; 2014; Neal, 2011), and island biogeography and parasite diversification (Mahrt, 1987; Perkins, 2001; Falk et al., 2015).
Malaria Parasites of Rodents
The rodent malaria parasites represent an unusual case where the parasite was first discovered in the insect host as opposed to the vertebrate one. In the 1940s entomological surveys in what is now the Democratic Republic of the Congo discovered a new species of Anopheles mosquito and some of them were found to contain sporozoites in their salivary glands. Given that this was long before DNA sequencing could be used to identify the hosts that they had fed on, assays that tested interactions with blood proteins were used, and rodents were identified as the likely source. A few years later, Grammomys surdaster (order Rodentia: family Muridae), the African woodland thicket rat were found infected with the parasites and the Plasmodium was successfully inoculated into white laboratory mice—and the rodent-malaria model system was born (Killick-Kendrick and Peters, 1978).
In a short span of time, 4 main species of rodent malaria were described and established as culture systems in laboratory mice. This model system played major roles in the early laboratory studies and characterization of malaria parasites, including early cell biology as well as genetic and immunological studies. The system was so important that the genome of a rodent malaria parasite, Plasmodium yoelii, was the very next to be sequenced following the publication of the P. falciparum genome (Carlton et al., 2002).
Malaria Parasites of Other Mammals
There are several other groups of mammals that are natural hosts for malaria parasites, including other primates, bats, and ungulates. However, generally these malaria parasites of other mammal groups have been less intensively studied than the model malaria parasites of humans and rodents.
Bats played an important role in the discovery of malaria parasites as it was Dionisi who first observed the cells in their blood as far back as 1898 (Perkins and Schaer, 2016). Given the dispersed nature of bats as hosts in the phylogeny of haemosporidians (Figure 2), they are also likely to be important transition hosts between bird hosts and other mammal hosts (Lutz et al., 2016b; Perkins and Schaer, 2016). Four primary genera have been found in various groups of bats worldwide. These include Plasmodium in Africa, Hepatocystis in Africa and Asia, Nycteria in Africa, and Polychromophilus from Africa, Europe, Central America, and South America. Several other monotypic genera (that is, a genus with a single species) have also been described from bat hosts, but their status will remain uncertain until genetic data can be collected. What was most interesting about the first molecular systematic studies of bat malaria is that they showed a very close relationship with the rodent malaria parasites that are so popular now as laboratory models (Schaer et al., 2013). The bat hosts of these parasites roost in trees that likely overlap ecologically with the arboreal thicket rats that serve as the natural hosts for the rodent-infecting Plasmodium species.
Several species of Plasmodium have been described from various ungulates including buffalo, goats, and small antelope. Plasmodium was also identified in a single white-tailed deer that had had its spleen removed in the southern United States (Garnham and Kuttler, 1980). Although deer are abundant in the eastern United States, the parasites in deer were not observed again until just recently, when several animals in Washington, DC, and other sites were shown to be infected by this parasite, now named P. odocoilei (Martinsen et al., 2016). Around the same time, other researchers also reported malaria parasites in hooved hosts ranging from goats in Africa to water buffalo in Thailand (Templeton et al., 2016). Phylogenetic analyses show that all ungulate malaria parasites discovered thus far are part of the same clade, but also that this clade is not truly part of the genus Plasmodium, but rather likely a distinct genus (Galen et al., 2018). The pathology of the white-tailed deer parasites and their tendency to infect and be virulent in very young animals is of interest to biomedical researchers, however, as although they are distantly related, this life history may mean that these parasites could serve as a model for P. falciparum infection in humans (Guggisberg et al., 2018; Perkins, 2018).
Malaria parasites have also been reported in 2 other groups of mammals: The colugos of Africa and elephant shrews of Malaysia (Perkins and Schaer, 2016). In both of these cases, there are many other species of haemosporidians known from the region, however, suggesting expanded host range. Nonetheless, the distribution of malaria parasites in mammals worldwide presents a puzzling pattern. There are no known haemosporidians from major groups of mammals such as carnivores or lagomorphs, and relatively few malaria parasite species have been described from the 2 largest orders of mammals (rodents and bats) and those that are known from these mammals have restricted geographic distributions.
Malaria Parasites of Humans
There are several species of Plasmodium that use humans as their hosts. In the past decade, many closely related lineages of parasites have been discovered to infect wild apes or other primates with a potential to also infect humans. The genetic divergences and host specificity among these novel ape malaria parasites are subjects of much study, thus it is likely imprudent to give an exact number of the taxa that do or could infect humans (McFadden, 2019). The 5 most common species that are found in humans are discussed below.
Plasmodium falciparum
Plasmodium falciparum, sometimes referred to as malignant tertian malaria, is the most virulent of the human-infecting species (the term tertian stems from the fact that the fevers from this infection become synchronized to every 2 days, and the Romans who first classified it as such did not use the concept of zero). It is widely distributed throughout the tropics, but is primarily concentrated in sub-Saharan Africa, Southeast Asia, and Oceania with known foci in South America. The reasons for the high virulence of this species are 2-fold. First, P. falciparum will invade any type of red blood cells and so can reach higher parasitemia than the other human parasites. Second, cells infected with P. falciparum become what might be thought of as sticky due to proteins expressed onto their surface as well as rigid and unable to bend, making it highly likely that they accumulate in capillaries causing a phenomenon known as sequestration. Sequestration in the brain or other vital tissues can cause death.
Plasmodium falciparum is transmitted among people primarily by the mosquito Anopheles gambiae, though many other species of Anopheles, such as A. arabiensis (Figure 4; see also Figure 5), are capable of transmission, depending on the geographic region (Molina-Cruz et al., 2016). In infected people the early trophozoite stages, which are called ring stages, are sometimes observed on thin blood smears but the mature stages are typically not observed due to the tendency of this species to sequester. Plasmodium falciparum is unusual amongst most mammal-infecting haemosporidians in that its gametocytes are crescent-shaped, rather than rounded (Figure 6A).
Virtually all human deaths attributed to malaria are caused by Plasmodium falciparum. It is currently present on all continents except for Europe (and Antarctica), but the largest proportion of fatalities is in children under 5 years-old who are living in sub-Saharan Africa. Because of its enormous global health importance, P. falciparum was the first malaria parasite—and one of the first organisms—to have its genome completely sequenced (Gardner et al., 2002).
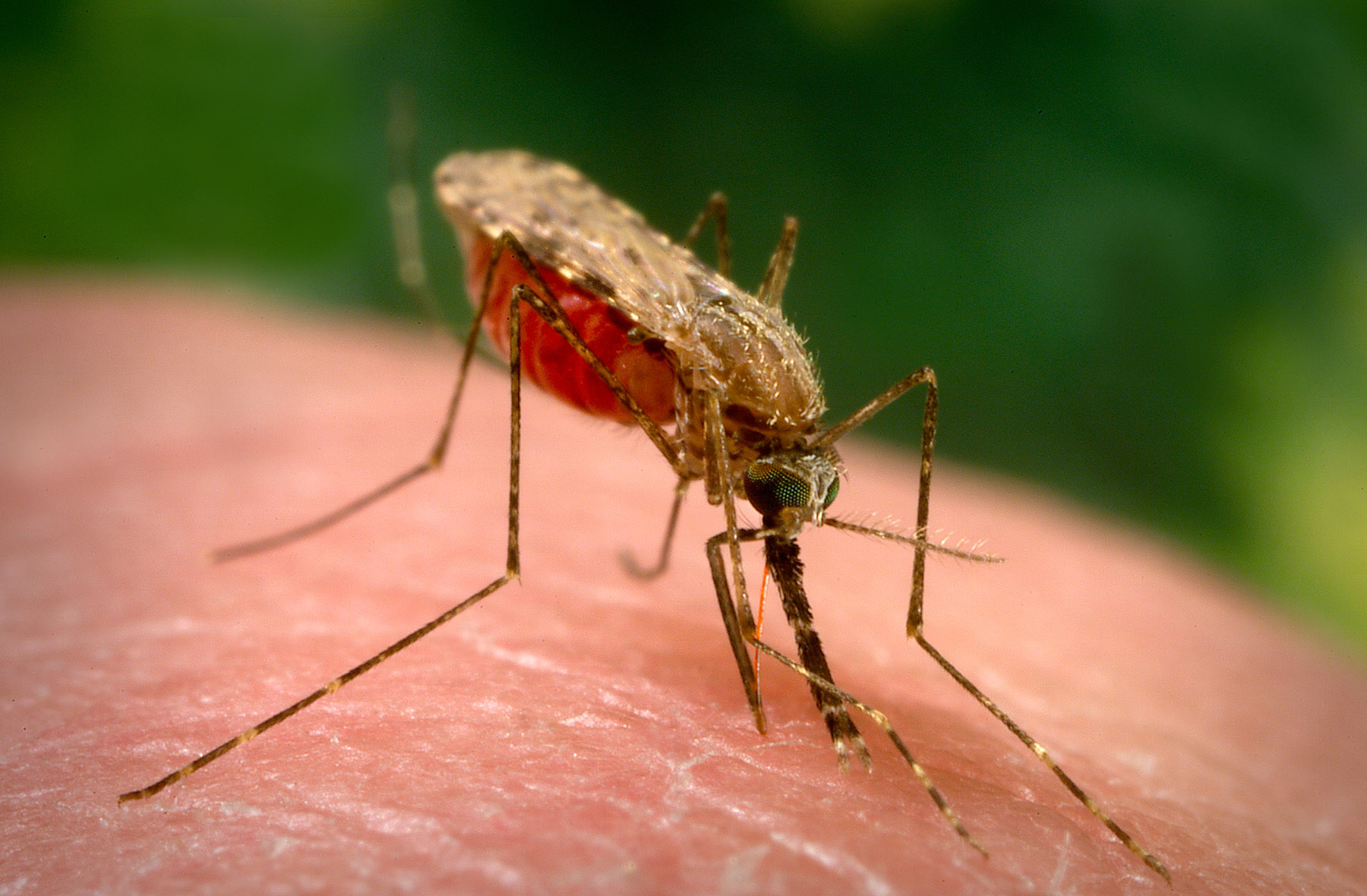
Figure 4. Anopheles arabiensis, one of the primary insect hosts of Plasmodium falciparum.
(Source: United States Centers for Disease Control and Prevention Public Health Image Library, image 18749; J. Gathany, 2014. Public domain.)
Figure 5. Mosquito morphology (female).
(Source: United States Centers for Disease Control and Prevention. Public domain.)
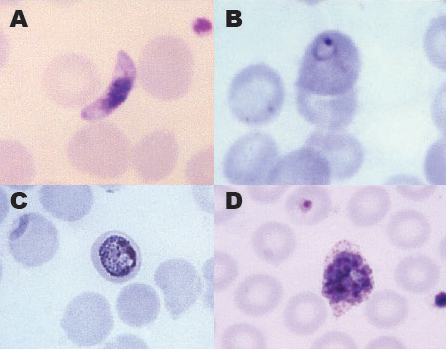
Figure 6. Stages of the 4 most common human malaria parasites. A) Gametocyte of Plasmodium falciparum. B) Ring stage of P. vivax. C) Trophozoite of P. malariae. D) Schizont of P. ovale.
(Source of photos: United States Centers for Disease Control and Prevention Public Health Image Library (A, image 4905; M. Melvin, 1966; C, image 5838; S. Glenn, 1979; D, image 5846; S. Glenn, 1979). Public domain.)
Plasmodium vivax
Plasmodium vivax, also known as benign tertian malaria, is also globally distributed and in the not-so-distant past, was even present in eastern cities of the United States such as Washington, DC, Philadelphia, and New York City. Because it is so geographically widespread, the economic burden of this parasite is very large—almost 3 billion people worldwide live in areas where P. vivax is present (Battle et al., 2012).
Although Plasmodium vivax is colloquially referred to as benign, it also has major health effects on its hosts, similar to those of P. falciparum, including anemia, jaundice, and even cerebral malaria (Bourgard et al., 2018). However, from an evolutionary standpoint P. vivax is more closely related to what are typically referred to as the macaque malarias, including P. cynomolgi and P. knowlesi (Galen et al., 2018). Unlike P. falciparum which is flexible in the blood cells it infects, P. vivax has a strong preference for using the reticulocytes of the host (Galinski and Barnwell, 1996; Galen et al., 2018) and so has a much lower parasitemia with only ring stages typically present in circulating blood (Figure 6B). It also produces the gametocyte stages much earlier in the vertebrate host, as early as 4 days even before clinical symptoms might present, a factor that could promote its transmission to new mosquitoes (Bourgard et al., 2018). What is perhaps most notable about P. vivax’s life cycle, however, is its presence of stages that remain viable in the liver called hypnozoites (Markus, 1980) that can later trigger a relapse in the disease.
Unlike Plasmodium falciparum, it has not been possible to culture P. vivax parasites in vitro in the laboratory, therefore it has been more challenging to work on this species. However, because of its great importance, the complete genome of P. vivax was one of the first malaria parasite genomes to be sequenced and was completed in 2008 (Carlton et al., 2008; Bourgard et al., 2018), opening up many new approaches to studying the biology of the parasite for its control.
Plasmodium malariae
Plasmodium malariae (Figure 4C), or benign quartan malaria has a 3-day periodicity (again, remember the lack of zero when it was given this name by the Romans). Recognized by the ancient Greeks, it was not until the late 19th century that Golgi made careful note that there seemed to be 2 parasites infecting people—1 with fevers every 48 hours (that is, tertian malaria) and 1 that had a slightly different periodicity, which he correlated with slight differences in the parasites that he observed in the patients’ blood (Garnham, 1966). Plasmodium malariae occurs throughout the world as well, but is most common in sub-Saharan Africa and the southwest Pacific though it can be very challenging to detect with just examination of blood films and thus molecular techniques such as PCR (polymerase chain reaction) are important to use (Garnham, 1966; Mueller et al., 2007).
In the early 1900s, a circus monkey was subjected to studies of its blood and a malaria parasite, similar in morphology to Plasmodium malariae, which was discovered and subsequently described as P. brasilianum (Garnham, 1966). These parasites were later observed in many wild monkeys in Central America and South America and for over a century were considered to be close relatives of—but not the same as—the human parasite. However, recent genetic results showed that these parasites were extremely similar (Fandeur et al., 2000), and in 2015, parasites that were genetically identical to P. brasilianum in wild howler monkeys were isolated from Indigenous Yanomami people living in Venezuela (Garnham, 1966; Lalremruata et al., 2015). When the complete genomes were sequenced, P. malariae and P. brasilianum were found to be the same species. A separate parasite, termed P. malariae-like, which was isolated from chimpanzees has since been found to be distinct (Rutledge et al., 2017).
Although Plasmodium malariae is typically considered a more benign form of malaria, it should not be dismissed as a public health concern. Because it can be difficult to diagnose with microscopy alone, it often goes undetected and may result in a fatal kidney disease (Eiam-Ong, 2003; Rutledge et al., 2017).
Plasmodium ovale
Until recently, Plasmodium ovale (Figure 6D) was generally considered to be the rarest form of the malaria parasites infecting humans. Plasmodium ovale has a rather scattered geographic distribution that primarily consists of western Africa, eastern Indonesia and New Guinea, and the Philippines, though of course due to the high mobility of humans, these parasites have also been reported in many other parts of the world (Mueller et al., 2007). Like P. malariae, P. ovale has also had a somewhat tumultuous taxonomic history. It was originally considered to be a variant of P. vivax, but was eventually described as a distinct species and named for the oval shape that some infected erythrocytes assume (Collins and Jeffery, 2005). Recently it was further split into 2 nominal subspecies on the basis of genetic data, P. o. wallikeri, and P. o. curtisi (see Sutherland et al., 2010).
Plasmodium knowlesi
In 2004, after a large number of malaria cases in Malaysian Borneo that were thought to have been Plasmodium malariae failed to amplify with species-specific primers, additional genetic testing confirmed that they were, in fact, naturally acquired infections of P. knowlesi, a parasite thought to be confined to macaques (Singh et al., 2004). Plasmodium knowlesi was later reported from mainland Malaysia as well as in isolated cases in Thailand and in China, though likely the latter was acquired in Myanmar (Singh et al., 2004; Cox-Singh et al., 2008). It has the shortest of periodicities of the human parasites, completing a cycle in just 24 hours and can reach extremely high, even fatal, human parasitemias and so proper diagnosis of this species and distinguishing it from P. malariae is very important (Cox-Singh et al., 2008).
Malaria Parasites in Apes
In the early part of the 20th century, parasitologists working in western Africa discovered 3 species of Plasmodium infecting both wild chimpanzees and gorillas. Although there were similarities to the species known to infect humans, distinct names were given to these taxa nonetheless. In 1 of these cases, P. reichenowi, genetic material was available as the parasite was isolated and cultured from a chimpanzee that had been imported into the United States. When molecular systematic analyses using parasite DNA were first attempted, the resulting phylogenetic trees supported the idea that P. reichenowi was closely related to P. falciparum, but nonetheless was a distinct species (Escalante et al., 1998; Perkins and Schall, 2001). However, the larger picture of malaria parasites in apes was largely unknown until around 2010. Understanding of malaria parasites in apes began to change during this period, as new samples were collected, first from captive apes and then via the screening of a large number of non-invasively collected ape fecal samples. These results showed that there were many genetically divergent malaria parasite lineages present in African chimpanzees and in western gorillas (though interestingly, never in bonobos nor eastern gorillas even though these host species were very well sampled; Liu et al., 2010). A total of 4 species of ape malaria parasite have now been named that appear to be close relatives. The phylogenetic relationships amongst the 6 species of Plasmodium (Laverania) suggest that these parasites have shifted among human, gorilla, and chimpanzee hosts several times, although previous transferal experiments had suggested that they were largely host specific. The possibility that wild ape malaria parasites might be able to jump into human hosts as zoonoses is worrisome not only to public health officials, who see the apes as a large host population that is not treatable and may represent a reservoir of the parasites, but also to conservation biologists, who are concerned that parasites that have undergone selective pressure in humans might be more virulent in the wild ape hosts.
Impact on Human Genetics
Because of the enormous impact on human health, it is not at all surprising that malaria has served as an important selective force throughout our history and in fact, the disease is thought to have been the strongest source of natural selection on human evolution at least in recent times (Kwiatkowski, 2005). Two prominent examples are often discussed, sickle cell anemia and Duffy coat receptors.
Sickle Cell Anemia
The primary molecule inside red blood cells—and in fact, the only significant protein inside mammalian red blood cells—is hemoglobin. This molecule is made up of 4 chains of amino acids with an iron group in the center. Hemoglobin is adept at binding to the 2 key molecules of aerobic respiration, oxygen and carbon dioxide, and serves as the transporter of these gases throughout the bloodstream of vertebrates. Mutations in the hemoglobin molecule have been identified that alter its function. One of these, known as HbS, can disrupt the structure of the red blood cell, making it fragile and likely to collapse into a sort of sickle shape as opposed to the normal round shape if the tension of the respiratory gases is abnormal.
If a person has 2 copies of the hemoglobin gene with this mutation, they will suffer from sickle cell anemia, a painful, largely untreatable, and sometimes fatal condition. One would predict, therefore, that natural selection would have removed these alleles from the human genome. And yet, they persist to this day. The reason is that people who are heterozygous for the variant hemoglobin gene have about a 10-fold higher protection from the forms of malaria infection most likely to cause death (Allison, 1954; Ackerman et al., 2005). If a red blood cell of a person who is heterozygous for sickle cell is infected with Plasmodium falciparum, the S type hemoglobin polymerizes, which then stalls the growth of the parasites (Archer et al., 2018).
Duffy coat Receptors
In human populations native to sub-Saharan Africa, there has been an almost complete fixation of a mutation that causes red blood cells to not express a protein that is necessary for the merozoite stage of Plasmodium vivax parasites to invade them (Kwiatkowski, 2005). This is known as the Duffy blood group-negative phenotype and makes those who have it essentially immune from P. vivax and P. knowlesi-caused forms of malaria.
Fighting Malaria as a Disease: Nets and Drugs
When Jesuit priests returned from missions in South America in the early 17th century, they brought bark from trees that indigenous Americans chewed to prevent shivering and other ailments, hypothesizing that it might also be helpful with the shivering that accompanied the fevers of malaria (Meshnick and Dobson, 2001). It did have some success in treating malaria patients (remember that at this point, they still did not know exactly how the disease was transmitted from person to person) and became widely known in Europe as the fever tree, Jesuits’ bark, or Peruvian bark. Around 1820, French chemists successfully extracted the main chemical component of the bark—quinine. Explorers searched the New World for trees that produced the highest concentrations of quinine and eventually seeds of Cinchona ledgeriana were used to start large plantations in Indonesia by the Dutch, and they controlled most of the world’s production of quinine. (An interesting side note is that because of its bitterness, it was often mixed with spirits to make it more palatable—this may have inspired the gin and tonic cocktail!)
Synthetic antimalarial drugs, particularly the drug chloroquine, became widely used especially in World War II. Chloroquine was very effective; it worked by disrupting the parasite’s ability to break down hemoglobin in the host cell and it was so widely used that in fact at one point it was sometimes mixed with table salt for mass distribution in malaria-endemic parts of the world. However, by the early 1960s Plasmodium falciparum parasites evolved resistance to chloroquine and the resistance quickly swept throughout most of the world. Mefloquine, sometimes known as Lariam®, and atovoquinone, often referred to as Malarone®, are 2 commonly used drugs that travelers to malarious parts of the world might be prescribed, though they are not safe to take for long periods of time (thus not usable for those people that live in malaria-endemic regions) and the parasites have evolved resistance to these compounds as well. In fact, there is not an anti-malarial drug that has been found or synthesized that the parasites have not evolved resistance to (Haldar et al., 2018). A compound known as qinghaosu or artemisinin (and its derivatives) that was used for centuries in China is now being produced and marketed, particularly in regions of Asia where parasites have evolved resistance to most of the other common anti-malarial compounds. It is a very effective drug and can even help patients in which the parasites have begun to sequester in capillaries. In most uses, it is administered along with other antimalarial drugs that have longer half lives in the body in an approach known as artemisin-combination therapy (White, 2008).

Figure 7. Children sleeping under a bed net. Ambitious programs to distribute insecticide-treated bed nets in malaria endemic areas has resulted in many fewer deaths in the past decade.
(Source: United States Global Health Initiative, 2006, https://commons.wikimedia.org/wiki/File:Malaria_prevention-Insecticide_treated_bed_net-PMI.jpg. Public domain.)
The other side of the malaria control coin is preventing people from acquiring the parasites in the first place, by stopping them from getting bitten by the mosquito vectors. This has been attempted through 3 main tactics: 1) Spraying insecticides, 2) poisoning the water sources where mosquito larvae are found, and 3) encouraging people, particularly children, to sleep under bed nets (Figure 7). All of these have had challenges or risks. In the 1950s there was a massive campaign to spray the insecticide DDT as a means of controlling mosquitoes and other insects. Although it worked very well to decrease mosquito populations, scientists quickly learned that this chemical exhibited bioaccumulation, or increased concentration as it moved up the food chain, ultimately being banned as a substance in the United States because of its severe environmental consequences. And, like the antimalarial drugs, the mosquitoes often quickly evolve resistance to these insecticides, causing them to lose their effectiveness (Hemingway et al., 2016). It is clear that combinations of methods and coordinated public health programs are going to have to be employed if malaria cases are going to be controlled, let alone if there is any hope of eradication of the disease. Some have argued that in order to be successful, evolutionary theory will need to be deployed into models of malaria control because these rapidly reproducing insects can more readily adapt to human-made chemicals than can be discovered and brought market (Read et al., 2009; Hemingway et al., 2016).
Chapter Reviewer
Ana Rivero, Maladies infectieuses et vecteurs: Écologie, génétique, evolution et contrôle, Institut de Recherche pour le Développement, Université de Montpellier, Montpellier, France
Literature Cited
Ackerman, H., S. Usen, M. Jallow, F. Sisay-Joof, et al. 2005. A comparison of case-control and family-based association methods: The example of sickle-cell and malaria. Annals of Human Genetics 69: 559–565. doi: 10.1111/j.1529-8817.2005.00180.x
Allison, A. C. 1954. Protection afforded by sickle-cell trait against subtertian malarial infection. British Medical Journal 1: 290–294. doi: 10.1136/bmj.1.4857.290
Archer, N. M., N. Petersen, M. A. Clark, C. O. Buckee, et al. 2018. Resistance to in sickle cell trait erythrocytes is driven by oxygen-dependent growth inhibition. Proceedings of the National Academy of Sciences of the United States of America 115: 7,350–7,355. doi: 10.1073/pnas.1804388115
Asghar, M., V. Palinauskas, N. Zaghdoudi-Allan, G. Valkiūnas, et al. 2016. Parallel telomere shortening in multiple body tissues owing to malaria infection. Proceedings of the Royal Society B: Biological Sciences 283: 20161184. doi: 10.1098/rspb.2016.1184
Asghar, M., V. Yman, M. V. Homann, K. Sondén, et al. 2017. Cellular aging dynamics after acute malaria infection: A 12-month longitudinal study. Aging Cell 17: e12702. doi: 10.1111/acel.12702
Atkinson, C. T., and M. D. Samuel. 2010. Avian malaria Plasmodium relictum in native Hawaiian forest birds: Epizootiology and demographic impacts on ‵apapane Himatione sanguinea. Journal of Avian Biology 41: 357–366. doi: 10.1111/j.1600-048X.2009.04915.x
Ayala, F. J., A. A. Escalante, and S. M. Rich. 1999. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia 41: 55–68.
Battle, K. E., P. W. Gething, I. R. F. Elyazar, C. L. Moyes, et al. 2012. The global public health significance of Plasmodium vivax. Advances in Parasitology 80: 1–111. doi: 10.1016/B978-0-12-397900-1.00001-3
Bensch, S., B. Canbäck, J. D. DeBarry, T. Johansson, et al. 2016. The genome of Haemoproteus tartakovskyi and its relationship to human malaria parasites. Genome Biology and Evolution 8: 1,361–1,373. doi: 10.1093/gbe/evw081
Bensch, S., O. Hellgren, and J. Pérez-Tris. 2009. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources 9: 1,353–1,358. doi: 10.1111/j.1755-0998.2009.02692.x
Bensch, S., J. Pérez-Tris, J. Waldenström, and O. Hellgren. 2004. Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: Multiple cases of cryptic speciation? Evolution 58: 1,617–1,621. doi: 10.1111/j.0014-3820.2004.tb01742.x
Böhme, U., T. D. Otto, J. A. Cotton, S. Steinbiss, et al. 2018. Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Research 28: 547–560. doi: 10.1101/gr.218123.116
Bourgard, C., L. Albrecht, C. A. Ana, P. Sunnerhagen, et al. 2018. Plasmodium vivax biology: Insights provided by genomics, transcriptomics and proteomics. Frontiers in Cellular and Infection Microbiology 8: 34. doi: 10.3389/fcimb.2018.00034
Carlton, J. M., J. H. Adams, J. C. Silva, S. L. Bidwell, et al. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757–763. doi: 10.1038/nature07327
Carlton, J. M., S. V. Angiuoli, B. B. Suh, T. W. Kooij, et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419: 512–519. doi: 10.1038/nature01099
Carter, R., and K. N. Mendis. 2002. Evolutionary and historical aspects of the burden of malaria. Clinical Microbiology Reviews 15: 564–594. doi: 10.1128/CMR.15.4.564-594.2002
Cibulskis, R. E., P. Alonso, J. Aponte, M. Aregawi, et al. 2016. Malaria: Global progress 2000–2015 and future challenges. Infectious Diseases of Poverty 5: 61. doi: 10.1186/s40249-016-0151-8
Collins, W. E., and G. M. Jeffery. 2005. Plasmodium ovale: Parasite and disease. Clinical Microbiology Reviews 18:570–581. doi: 10.1128/CMR.18.3.570-581.2005
Cox-Singh, J., T. M. E. Davis, K.-S. Lee, S. S. G. Shamsul, et al. 2008. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clinical Infectious Diseases 46: 165–171. doi: 10.1086/524888
Eiam-Ong, S. 2003. Malarial nephropathy. Seminars in Nephrology 23: 21–33. doi: 10.1053/snep.2003.50002
Ellis, V. A., M. D. Collins, M. C. I. Medeiros, E. H. R. Sari, et al. 2015. Local host specialization, host-switching, and dispersal shape the regional distributions of avian haemosporidian parasites. Proceedings of the National Academy of Sciences of the United States of America 112: 11,294–11,299. doi: 10.1073/pnas.1515309112
Escalante, A. A., D. E. Freeland, W. E. Collins, and A. A. Lal. 1998. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proceedings of the National Academy of Sciences of the United States of America 95: 8,124–8,129. doi: 10.1073/pnas.95.14.8124
Falk, B. G., R. E. Glor, and S. L. Perkins. 2015. Clonal reproduction shapes evolution in the lizard malaria parasite Plasmodium floridense. Evolution 69: 1,584–1,596. doi: 10.1111/evo.12683
Fandeur, T., B. Volney, C. Peneau, and B. de Thoisy. 2000. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology 120: 11–21. doi: 10.1017/S0031182099005168
Galen, S. C., J. Borner, E. S. Martinsen, J. Schaer, et al. 2018. The polyphyly of Plasmodium: Comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict. Royal Society Open Science 5: 171780. doi: 10.1098/rsos.171780
Galinski, M. R., and J. W. Barnwell. 1996. Plasmodium vivax: Merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitology Today 12: 20–29. doi: 10.1016/0169-4758(96)80641-7
Gardner, M. J., N. Hall, E. Fung, O. White, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. doi: 10.1038/nature01097
Garnham, P. C. C. 1966. Malaria Parasites and Other Haemosporidia. Blackwell Scientific, Oxford, United Kingdom, 1,114 p.
Garnham, P. C. C., and K. L. Kuttler. 1980. A malaria parasite of the white-tailed deer (Odocoileus virginianus) and its relation with known species of Plasmodium in other ungulates. Proceedings of the Royal Society B: Biological Sciences 206: 395–402. doi: 10.1098/rspb.1980.0003
Guggisberg, A. M., K. A. Sayler, S. M. Wisely, and A. R. Odom John. 2018. Natural history of malaria infection in farmed white-tailed deer. mSphere 3: e00067-18. doi: 10.1128/mSphere.00067-18
Haldar, K., S. Bhattacharjee, and I. Safeukui. 2018. Drug resistance in Plasmodium. Nature Reviews Microbiology 16: 156–170. doi: 10.1038/nrmicro.2017.161
Hemingway, J., H. Ranson, A. Magill, J. Kolaczinski, et al. 2016. Averting a malaria disaster: Will insecticide resistance derail malaria control? Lancet 387: 1,785–1,788. doi: 10.1016/S0140-6736(15)00417-1
Huff, C. G., and F. Coulston. 1946. The relation of natural and acquired immunity of various avian hosts to the cryptozoites and metacryptozoites of Plasmodium gallinaceum and Plasmodium relictum. Journal of Infectious Diseases 78: 99–117. doi: 10.1093/infdis/78.2.99
Killick-Kendrick, R., and W. Peters. 1978. Rodent Malaria. Academic Press, New York, New York, United States, 406 p.
Kwiatkowski, D. P. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. American Journal of Human Genetics 77: 171–192. doi: 10.1086/432519
Lalremruata, A., M. Magris, S. Vivas-Martínez, M. Koehler, et al. 2015. Natural infection of Plasmodium brasilianum in humans: Man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine 2: 1,186–1,192. doi: 10.1016/j.ebiom.2015.07.033
Levine, N. D. 1988. The Protozoan Phylum Apicomplexa, Volume 2. CRC Press, Boca Raton, Florida, United States, 154 p.
Lutz, H. L., N. J. Marra, F. Grewe, J. S. Carlson, et al. 2016a. Laser capture microdissection microscopy and genome sequencing of the avian malaria parasite, Plasmodium relictum. Parasitology Research 115: 4,503–4,510. doi: 10.1007/s00436-016-5237-5
Lutz, H. L., B. D. Patterson, J. C. Kerbis Peterhans, W. T. Stanley, et al. 2016b. Diverse sampling of East African haemosporidians reveals chiropteran origin of malaria parasites in primates and rodents. Molecular Phylogenetics and Evolution 99: 7–15. doi: 10.1016/j.ympev.2016.03.004
Mahrt, J. L. 1987. Lizard malaria in Arizona: Island biogeography of Plasmodium chiricahuae and Sceloporus jarrovi. Southwestern Naturalist 32: 347. doi: 10.2307/3671451
Markus, M. 1980. The malarial hypnozoite. Lancet 315: 936. doi: 10.1016/s0140-6736(80)90871-5
Martínez-de la Puente, J., J. Martínez, J. Rivero-de Aguilar, J. Herrero, et al. 2011. On the specificity of avian blood parasites: Revealing specific and generalist relationships between haemosporidians and biting midges. Molecular Ecology 20: 3,275–3,287. doi: 10.1111/j.1365-294X.2011.05136.x
Martinsen, E. S., N. McInerney, H. Brightman, K. Ferebee, et al. 2016. Hidden in plain sight: Cryptic and endemic malaria parasites in North American white-tailed deer (Odocoileus virginianus). Science Advances 2: e1501486. doi: 10.1126/sciadv.1501486
McFadden, G. I. 2019. Plasmodium: More don’ts. Trends in Parasitology 35: 4–6. doi: 10.1016/j.pt.2018.10.002
Meshnick, S. R., and M. J. Dobson. 2001. The history of antimalarial drugs. In P. J. Rosenthal, ed. Antimalarial Chemotherapy: Mechanisms of Action, Resistance, and New Directions in Drug Discovery. Springer, New York, New York, United States, p. 15–25.
Molina-Cruz, A., M. M. Zilversmit, D. E. Neafsey, D. L. Hartl, et al. 2016. Mosquito vectors and the globalization of Plasmodium falciparum malaria. Annual Review of Genetics 50: 447–465. doi: 10.1146/annurev-genet-120215-035211
Mueller, I., P. A. Zimmerman, and J. C. Reeder. 2007. Plasmodium malariae and Plasmodium ovale: The “bashful” malaria parasites. Trends in Parasitology 23: 278–283. doi: 10.1016/j.pt.2007.04.009
Neal, A. T. 2011. Male gametocyte fecundity and sex ratio of a malaria parasite, Plasmodium mexicanum. Parasitology 138: 1,203–1,210. doi: 10.1017/S0031182011000941
Neal, A. T., and J. J. Schall. 2010. Gametocyte sex ratio in single-clone infections of the malaria parasite Plasmodium mexicanum. Parasitology 137: 1,851–1,859. doi: 10.1017/S0031182010000909
Neal, A. T., and J. J. Schall. 2014. Testing sex ratio theory with the malaria parasite Plasmodium mexicanum in natural and experimental infections. Evolution 68: 1,071–1,081. doi: 10.1111/evo.12334
Osgood, S. M., and J. J. Schall. 2004. Gametocyte sex ratio of a malaria parasite: Response to experimental manipulation of parasite clonal diversity. Parasitology 128: 23–29. doi: 10.1017/S0031182003004207
Palinauskas, V., R. Žiegytė, J. Šengaut, and R. Bernotienė. 2018. Different paths, the same virulence: Experimental study on avian single and co-infections with Plasmodium relictum and Plasmodium elongatum. International Journal for Parasitology 48: 1,089–1,096. doi: 10.1016/j.ijpara.2018.08.003
Perkins, S. L. 2018. Malaria in farmed ungulates: An exciting new system for comparative parasitology. mSphere 3: e00161-18. doi: 10.1128/mSphere.00161-18
Perkins, S. L. 2001. Phylogeography of Caribbean lizard malaria: Tracing the history of vector borne parasites. Journal of Evolutionary Biology 14: 34–45. doi: 10.1046/j.1420-9101.2001.00261.x
Perkins, S. L., and J. Schaer. 2016. A modern menagerie of mammalian malaria. Trends in Parasitology 32: 772–782. doi: 10.1016/j.pt.2016.06.001
Perkins, S. L., and J. J. Schall. 2002. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. Journal of Parasitology 88: 972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2
Pigeault, R., A. Nicot, S. Gandon, and A. Rivero. 2015. Mosquito age and avian malaria infection. Malaria Journal 14: 383. doi: 10.1186/s12936-015-0912-z
Read, A. F., P. A. Lynch, and M. B. Thomas. 2009. How to make evolution-proof insecticides for malaria control. PLoS Biology 7: e1000058. doi: 10.1371/journal.pbio.1000058
Remot, F., V. Ronget, H. Froy, B. Rey, et al. 2022. Decline in telomere length with increasing age across nonhuman vertebrates: A meta‐analysis. Molecular Ecology 31: 5,917–5,932. doi: 10.1111/mec.16145
Rivero, A., and S. Gandon. 2018. Evolutionary ecology of avian malaria: Past to present. Trends in Parasitology 34: 712–726. doi: 10.1016/j.pt.2018.06.002
Rutledge, G. G., U. Böhme, M. Sanders, A. J. Reid, et al. 2017. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution [Letter]. Nature 542: 101–104. doi: 10.1038/nature21038
Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415: 680–685. doi: 10.1038/415680a
Samuel, M. D., P. H. F. Hobbelen, F. DeCastro, J. A. Ahumada, et al. 2011. The dynamics, transmission, and population impacts of avian malaria in native Hawaiian birds: A modeling approach. Ecological Applications 21: 2,960–2,973. doi: 10.1890/10-1311.1
Schaer, J., S. L. Perkins, J. Decher, F. H. Leendertz, et al. 2013. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proceedings of the National Academy of Sciences of the United States of America 110: 17,415–17,419. doi: 10.1073/pnas.1311016110
Schall, J. J. 2009. Do malaria parasites follow the algebra of sex ratio theory? Trends in Parasitology 25: 120–123. doi: 10.1016/j.pt.2008.12.006
Schall, J. J. 1990. The ecology of lizard malaria. Parasitology Today 6: 264–269. doi: 10.1016/0169-4758(90)90188-A
Schall, J. J. 1983. Lizard malaria: Cost to vertebrate host’s reproductive success. Parasitology 87: 1. doi: 10.1017/S0031182000052367
Schall, J. J. 2000. Transmission success of the malaria parasite Plasmodium mexicanum into its vector: Role of gametocyte density and sex ratio. Parasitology 121: 575–580. doi: 10.1017/S0031182000006818
Schall, J. J., and C. M. Staats. 1997. Parasites and the evolution of extravagant male characters: Anolis lizards on Caribbean islands as a test of the Hamilton-Zuk hypothesis. Oecologia 111: 543–548. doi: 10.1007/s004420050269
Schall, J. J., and C. M. Staats. 2002. Virulence of lizard malaria: Three species of Plasmodium infecting Anolis sabanus, the endemic anole of Saba, Netherlands Antilles. Copeia 2002: 39–43. doi: 10.1643/0045-8511(2002)002[0039:VOLMTS]2.0.CO;2
Singh, B., L. Kim Sung, A. Matusop, A. Radhakrishnan, et al. 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363: 1,017–1,024. doi: 10.1016/S0140-6736(04)15836-4
Sutherland, C. J., N. Tanomsing, D. Nolder, M. Oguike, et al. 2010. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. Journal of Infectious Diseases 201: 1,544–1,550. doi: 10.1086/652240
Svensson-Coelho, M., V. A. Ellis, B. A. Loiselle, J. G. Blake, et al. 2014. Reciprocal specialization in multihost malaria parasite communities of birds: A temperate-tropical comparison. American Naturalist 184: 624–635. doi: 10.1086/678126
Telford, S. R. 2008. Hemoparasites of the Reptilia: Color Atlas and Text, 1st edition. CRC Press, Boca Raton, Florida, United States, 376 p.
Templeton, T. J., E. Martinsen, M. Kaewthamasorn, and O. Kaneko. 2016. The rediscovery of malaria parasites of ungulates. Parasitology 143: 1,501–1,508. doi: 10.1017/S0031182016001141
Tonkin, I. M., and F. Hawking. 1947. The technique of testing chemotherapeutic action on Plasmodium gallinaceum. British Journal of Pharmacology and Chemotherapy 2: 221–233. doi: 10.1111/j.1476-5381.1947.tb00339.x
Valkiūnas, G. 2004. Avian Malaria Parasites and other Haemosporidia, 1st edition. CRC Press, Boca Raton, Florida, United States, 946 p.
Valkiūnas, G., M. Ilgūnas, D. Bukauskaitė, K. Fragner, et al. 2018. Characterization of Plasmodium relictum, a cosmopolitan agent of avian malaria. Malaria Journal 17: 184. doi: 10.1186/s12936-018-2325-2
van Riper, C., S. G. van Riper, M. Lee Goff, and M. Laird. 1986. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecological Monographs 56: 327–344. doi: 10.2307/1942550
Vézilier, J., A. Nicot, S. Gandon, and A. Rivero. 2012. Plasmodium infection decreases fecundity and increases survival of mosquitoes. Proceedings of the Royal Society B: Biological Sciences 279: 4,033–4,041. doi: 10.1098/rspb.2012.1394
Videvall, E., C. K. Cornwallis, V. Palinauskas, G. Valkiūnas, et al. 2015. The avian transcriptome response to malaria infection. Molecular Biology and Evolution 32: 1,255–1,267. doi: 10.1093/molbev/msv016
Waters, A. P., D. G. Higgins, and T. F. McCutchan. 1991. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proceedings of the National Academy of Sciences of the United States of America 88: 3,140–3,144. doi: 10.1073/pnas.88.8.3140
Weinberg, J., J. T. Field, M. Ilgūnas, D. Bukauskaitė, et al. 2018. De novo transcriptome assembly and preliminary analyses of two avian malaria parasites, Plasmodium delichoni and Plasmodium homocircumflexum. Genomics S0888-7543: 30431-2. doi: 10.1016/j.ygeno.2018.12.004
White, N. J. 2008. Qinghaosu (artemisinin): The price of success. Science 320: 330–334. doi: 10.1126/science.1155165
WHO (World Health Organization). 2023. Malaria vaccine plays critical role in turning the tide on malaria in Ghana. World Health Organization, Geneva, Switzerland. https://www.afro.who.int/countries/ghana/news/malaria-vaccine-plays-critical-role-turning-tide-malaria-ghana
WHO (World Health Organization). 2021. World malaria report 2021. World Health Organization, Geneva, Switzerland, 263 p.
Yan, J., J. Martínez-de la Puente, L. Gangoso, R. Gutiérrez-López, et al. 2018. Avian malaria infection intensity influences mosquito feeding patterns. International Journal for Parasitology 48: 257–264. doi: 10.1016/j.ijpara.2017.09.005
Supplemental Reading
Ayala, F. J., and W. M. Fitch. 1992. Phylogeny of Plasmodium falciparum. Parasitology Today 8: 74–75. doi: 10.1016/0169-4758(92)90234-s
Borner, J., C. Pick, J. Thiede, O. M. Kolawole, et al. 2016. Phylogeny of haemosporidian blood parasites revealed by a multigene approach. Molecular Phylogenetics and Evolution 94: 221–231. doi: 10.1016/j.ympev.2015.09.003
Cornejo, O. E., and A. A. Escalante. 2006. The origin and age of Plasmodium vivax. Trends in Parasitology 22: 558–563. doi: 10.1016/j.pt.2006.09.007
Escalante, A. A., and F. J. Ayala. 1995. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proceedings of the National Academy of Sciences of the United States of America 92: 5,793-5,797. doi: 10.1073/pnas.92.13.5793
Escalante A. A., and F. J. Ayala. 1994. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proceedings of the National Academy of Sciences of the United States of America 91: 11,373-11,377. doi: 10.1073/pnas.91.24.1137
Escalante, A. A., A. S. Cepeda, and M. A. Pacheco. 2022. Why Plasmodium vivax and Plasmodium falciparum are so different? A tale of two clades and their species diversities. Malaria Journal 21: 139. doi: 10.1186/s12936-022-04130-9
Escalante, A. A., O. E. Cornejo, D. E. Freeland, A. C. Poe, et al. 2005. A monkey’s tale: The origin of Plasmodium vivax as a human malaria parasite. Proceedings of the National Academy of Sciences of the United States of America 102: 1,980–1,985. doi: 10.1073/pnas.0409652102
Escalante, A. A., O. E. Cornejo, A. Rojas, V. Udhayakumar, et al. 2004. Assessing the effect of natural selection in malaria parasites. Trends in Parasitology 20: 388–395. doi: 10.1016/j.pt.2004.06.002
Hayakawa, T., R. Culleton, H. Otani, T. Horii, et al. 2008. Big bang in the evolution of extant malaria parasites. Molecular Biology and Evolution 25: 2,233–2,239. doi: 10.1093/molbev/msn171
Martinsen, E. S., S. L. Perkins, and J. J. Schall. 2008. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Molecular Phylogenetics and Evolution 47: 261–273 doi: 10.1016/j.ympev.2007.11.012
Neafsey, D. E., K. Galinsky, R. H. Jiang, L. Young, et al. 2012. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nature Genetics 44: 1,046–1,050. doi: 10.1038/ng.2373
Outlaw, D. C., and R. E. Ricklefs. 2011. Rerooting the evolutionary tree of malaria parasites. Proceedings of the National Academy of Sciences of the United States of America 108: 13,183–13,187. doi: 10.1073/pnas.1109153108
Pacheco, M. A., L. M. P. Ceríaco, N. E. Matta, M. Vargas-Ramírez, et al. 2020. A phylogenetic study of Haemocystidium parasites and other Haemosporida using complete mitochondrial genome sequences. Infection, Genetics, and Evolution 85: 104576. doi: 10.1016/j.meegid.2020.104576
Pacheco, M. A., M. López-Pérez, A. F. Vallejo, S. Herrera, et al. Multiplicity of infection and disease severity in Plasmodium vivax. PLoS Neglected Tropical Diseases 10: e0004355. doi: 10.1371/journal.pntd.0004355
Perkins S. L. 2008. Molecular systematics of the three mitochondrial protein-coding genes of malaria parasites: Corroborative and new evidence for the origins of human malaria. Mitochondrial DNA 19: 471-478. doi: 10.1080/19401730802570926
Ramiro, R. S., S. E. Reece, and D. J. Obbard. 2012. Molecular evolution and phylogenetics of rodent malaria parasites. BMC Evolutionary Biology 12: 219. doi: 10.1186/1471-2148-12-219
Ricklefs R. E., and D. C. Outlaw. 2010. A molecular clock for malaria parasites. Science 329: 226–229. doi: 10.1126/science.1188954
Rodrigues, P. T., H. O. Valdivia, T. C. de Oliveira, J. M. P. Alves, et al. 2018. Human migration and the spread of malaria parasites to the new World. Scientific Reports 8: 1993. doi: 10.1038/s41598-018-19554-0
Silva, J. C., A. Egan, C. Arze, J. L. Spouge, et al. 2015. A new method for estimating species age supports the coexistence of malaria parasites and their mammalian hosts. Molecular Biology and Evolution 32: 1,354–1,364. doi: 10.1093/molbev/msv005
Torres, K., M. U. Ferreira, M. C. Castro, A. A. Escalante, et al. Malaria resilience in South America: Epidemiology, vector biology, and immunology insights from the Amazonian International Center of Excellence in Malaria Research Network in Peru and Brazil. American Journal of Tropical Medicine and Hygiene 107 (Supplement): 168–181. doi: 10.4269/ajtmh.22-0127
Waters, A. P., D. G. Higgins, and T. F. McCutchan. 1991. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proceedings of the National Academy of Sciences of the United States of America 88: 3,140–3,144. doi: 10.1073/pnas.88.8.3140
Definition: A parasite that cannot exist without a host during all or some portion of the life cycle. see facultative parasite
Adjective
From Greek: heteros = different; xenos = host/guest/stranger
Definition: Having more than one host during a parasite's life cycle
Noun
From Greek: spora = seed; zoon = animal
Definition: The stage of development of a sporoblast which has divided and exited the oocyst into the hemocoel and migration begins; the malarial stage found in the salivary glands of insects
Definition: Any method of reproduction not involving fertilization, as that by fission, fragmentation, spore production, budding, vegetative reproduction, and gemmule formation
Definition: The two cells resulting from division of a single cell
Noun
From Greek: gamete = wife; kytos = container
Definition 1: A spermatocyte or oocyte
Definition 2: Sexual stage of the malarial parasite in the blood which upon being taken into the mosquito host may produce gametes
Noun
From Greek: oion = egg; kinetos = move
Definition: Among insects, a motile, elongate zygote of a Plasmodium that encysts in the stomach wall of a Culicidae
Definition: Host in which the terminal (frequently sexual) stage of the parasite occurs
Synonym: Primary host
Definition: One which alternates with the definitive host in which the parasite passes through partial development, but not to sexual maturity
Noun
From Latin: vehere = to carry
Definition 1: Any carrier, particularly an animal, that transmits a disease organism from one host to another
Defintion 2: In helminthic disease, an intermediate host that seeks out the definitive host; such as a mosquito

