11 Trypanosoma (Genus)
Ana Maria Jansen; Samanta C. Chagas Xavier; and André Luiz Rodrigues Roque
Phylum Euglenozoa
Class Kinetoplastea
Order Trypanosomatida
Family Trypanosomatidae
Genus Trypanosoma
Introduction
It is difficult to know exactly where to begin the introduction of the flagellated eukaryotic parasites that are classified in the genus Trypanosoma. This is because there is so much that can be learned about parasitism from the study of the morphological characters of these organisms; or by studies of how their populations cycle through individual mammals; or how they grow and reproduce in their insect vectors; or how these parasites are maintained in populations of mammals and their arthropod vectors; there is even a species that jumps from host to host without the inconvenience of having to use a vector; it has evolved past the need for a vector and instead uses the behavior of its infected host to transfer to a new potential host. However, starting off with a general classification of the group is generally a good idea, so that students who are just beginning, or even an advanced student, can see where the parasite belongs in the general scheme of life.
The trypanosomes are a monophyletic group (meaning, having a common origin from a single ancestral species) of single-celled eukaryotes (eu = true, karyon = nucleus; Greek), or those organisms that have a true nucleus. Obligate parasites of the genus Trypanosoma are included within the superorder Kinetoplastida. One of the current classifications looks like this:
Domain Eukaryota
Phylum Euglenozoa
Class Kinetoplastida
Order Trypanosomatida
Family Trypanosomatidae
Genus Trypanosoma
As seen in the list above, all known trypanosomatids are classified in the order Trypanosomatida, family Trypanosomatidae (see Moreira et al., 2004).
Members of this unique family are characterized by the single celled organism having an elongated cell body containing one flagellum emerging at the anterior part of the parasite and a single mitochondrion that is distributed throughout the cell body (see Figure 1 for details about the general morphology). The compressed DNA of this mitochondrion is called a kinetoplast, which is composed of maxi-circles and mini-circles, and is situated close to the base of the flagellar pocket (the place in the body from which the flagellum originates). Maxi-circles encode the genes required to perform the physiological function of oxidative phosphorylation that occurs, for example, in the midgut of the tsetse fly, and mini-circles are responsible for mRNA editing (Shlomai, 2004).
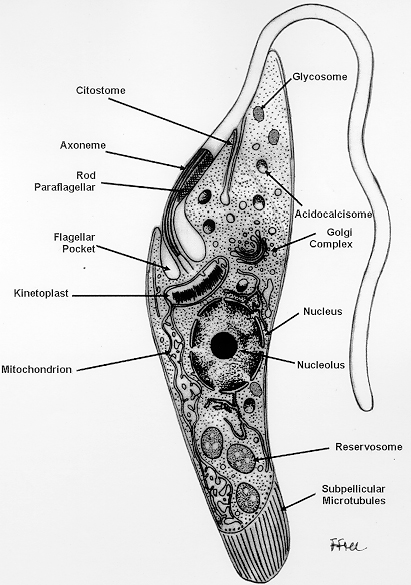
Figure 1. Schematic drawing, based on information obtained with the transmission electron microscope, showing the various structures found in the epimastigote form of Trypanosoma cruzi.
(Source of diagram: Souza, 1999. License: CC BY 4.0.)
While species included in the family Trypanosomatidae have variable and distinct life histories, all species are obligate parasites. This family includes predominantly monoxenic forms (meaning, living on one host species), but also includes 4 genera with heteroxenic species (meaning, living in more than one host species) that are parasites of: 1) Plants (Phytomonas spp.); 2) sloths (Endotrypanum spp.); 3) other mammals and lizards (Leishmania spp.); and 4) all vertebrate classes (Trypanosoma spp.). Relatively recently, a fifth genus was proposed (Porcisia sp.) to include Neotropical porcupine-infecting species, previously known as Leishmania hertigi and L. deane (see Espinosa et al., 2018).
Concerning the monoxenic trypanosomatids, currently from 14 to 17 genera of parasites have been recognized, including those parasitizing Diptera, Hymenoptera, Siphonaptera, and Hemiptera. These monoxenic forms are distributed among the genera: Angomonas, Blechomonas, Blastocrithidia, Crithidia, Herpetomonas, Kentomonas, Leptomonas, Lotmaria, Novymonas, Paratrypanosoma, Sergeia, Strigomonas, Wallacemonas, and Zelonia (see Kaufer et al., 2017), as well as Jaeinimonas, Lafontella, and Rhynchoidomonas (see D’Ávila-Levy et al., 2015). Although described as monoxenic insect parasites, this trait (occurring in insects) seems not to be strict, since some of them have been reported parasitizing mammals, probably after being transmitted by insects. In fact, co-infections of Leptomonas seymouri and Leishmania donovani have been reported in patients with visceral leishmaniasis (Ghosh et al., 2012), and records of concomitant infections by Leptomonas species and Herpetomonas samuelpessoai in a HIV-positive human patient (Morio et al., 2008). In addition, there are records of Herpetomonas species infecting plants (Borghesan et al., 2013), and reports of Blastocrithidia species and Chritidia mellificae occurring in bats (Hodo et al., 2016; Rangel et al., unpublished data). Interestingly, when experimentally injected in the scent glands of opossums, Leptomonas species and Chritidia speecies not only established the infection, but also multiplied. The parasitism of scent glands of Didelphis species by monoxenic trypanosomatids was interpreted by Deane and colleagues (1984) as being the stepping-stone of trypanosomatids on their way to adapt to a mammal host.
Species of the genus Trypanosoma are parasites of vertebrates that are, with a few exceptions, transmitted among vertebrates by invertebrate vector hosts (Figure 2). Four primary morphological stages are recognized among trypanosomes: The extracellular trypomastigote, epimastigote, and spheromastiogote forms, and the intracellular amastigote form. These morphological forms occur in various stages among trypanosomatids with different species sometimes manifesting these forms differently. In 1972, the British protozoologist Cecil Hoare, in his foundational monograph, proposed the separation of the genus Trypanosoma in 2 sections based on the transmission mode of these parasites: Stercoraria and Salivaria (see Hoare, 1972). The divergence between these 2 sections would have occurred in the Cretaceous period (approximately 100 Ma = million years ago), accompanying the breakup of Gondwanaland and the separation of Africa from South America, Antarctica, and Australia. According to this classification that was widely accepted and used up to the current time, there are 3 sub-genera in Stercoraria section: T. (Herpetosoma) sp., T. (Megatrypanum) sp., and T. (Schizotrypanum) sp.; and 4 in Salivaria section: T. (Dutonella) sp., T. (Nannomonas) sp., T. (Trypanozon) sp., and T. (Pycnomonas). An eighth subgenus, Tejeraia, was proposed at a later date to include Trypanosoma rangeli, a South American parasite previously included in the Herpetosoma subgenus, but, as it later became clear, it is in fact transmitted by several hematophagous triatomine species (Añez, 1982).
The recent molecular tools with their great discrimination power that have been developed in the last decades have resulted in extensive revisions and descriptions of new genera and species. In fact, classifications up to 3 decades ago were made based on parasite morphology combined with the host species in which trypanosomatids were found. Another important point is the attention that was classically given to trypanosomatids with enzootic potential or potential to affect animals of economic interest. The current awareness of the importance of biodiversity (and parasites as important components of this) has widened the focus of attention in order to increase the interest in parasites not necessarily of humans or of domestic animals (Gardner and Campbell, 1992; Poulin, 2014; Jenkins et al., 2015).
Section Salivaria
Trypanosomes included in this section are highly prevalent in sub-Saharan Africa in the so termed Tsetse-Belt region and represent important health threats for humans and other animals with concomitant economic impacts in the areas of occurrence. Several species of the section Salivaria occur and can be found being transmitted in several countries beyond the African continent and the possibility of even greater geographic dispersion should not be neglected (Osório et al., 2008; Aregawi et al., 2019). Transmission occurs through inoculation of infective metacyclic forms as the insect vector feeds on blood of an infected mammal. For most species of salivarian trypanosomes but one, the only proven insect-vectors are various species of the hematophagous flies in the genus Glossina (the infamous and notorious tsetse flies of Africa). The only species that can be transmitted mechanically by other dipteran is T. vivax. Glossina species (Diptera: Glossinidae) (Figures 3 and 4) are the biological vectors of all salivarian trypanosomes and an individual fly may harbor more than one trypanosome species (Van den Bossche et al., 2004). All of the approximately 33 known species of Glossina that have been tested are able to act as vectors of salivarian trypanosomes; However, in contrast to many species of dipteran vectors, such as the blood-feeding Tabanidae, in which only females take blood meals, both sexes of Glossina are hematophagous and are capable of transmitting trypanosomes via saliva during blood feeding. Moreover, Trypanosoma vivax (another species of Salivarian trypanosome that lives in African mammals) can be transmitted by Glossina as well as by other hematophagous diptera (Fetene et al., 2021). Interestingly, Trypanosoma evansi does not appear to be transmitted by Glossina species and is mechanically transmitted from ungulate to ungulate by tabanids (order Diptera: family Tabanidae) and other hematophagous vector insects such as species of Stomoxys, Atylotus, Lyperosia (see Brun et al., 1998), and other slash and bite blood feeders such as Desmodus rotundus (common vampire bat) or Diaemus youngi the white-winged vampire bat in the Americas. Trypanosoma equiperdum the causative agent of Dourine in horses is a venereally-transmitted trypanosome and is generally considered to have a cosmopolitan (worldwide) distribution with the parasite generally absent in North America (north of Mexico), Australia, and western Europe (Brun et al., 1998; Gizaw et al., 2017).
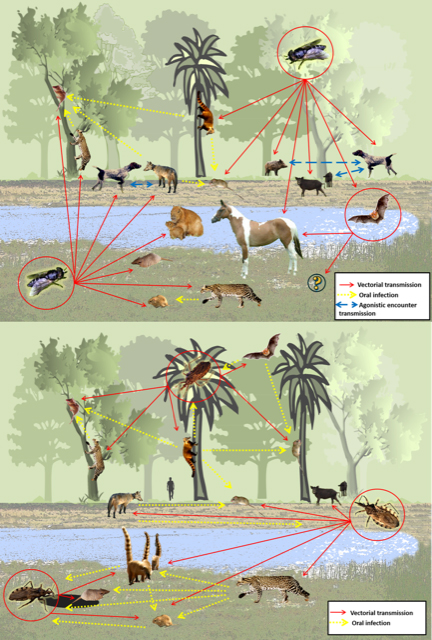
Figure 2. The wild and synanthropic reservoirs of Leishmania species represented graphically.
(Source: Rodrigues Roque and Jansen, 2014. License: CC BY-NC-SA 4.0 International.)
The development of salivarian trypanosomes in the tsetse fly may be generally complex among Trypanosoma species and may be influenced by the insect immune response modulated by the fly’s gut and symbiotic microbiota. Sexual reproduction in trypaonsomes has been reported, but only when the protozoans are actively reproducing within the insect vector-hosts (Gibson, 2015). Three interaction patterns of trypanosomes in the tsetse fly are recognized:
1) Trypanosoma vivax group (Dutonella subgenus) includes the trypanosomes with the lowest degree of interaction with the insect vector. This species is found across the Tsetse-Belt in Africa as well as in several countries in Latin America, occurring in wild and domestic animals. Recorded hosts for T. vivax include water buffalo, cattle, dogs, dromedary camels, horses, suids, and small ruminants. As noted, this species also occurs in wild animals that can serve as reserviors of infection for domestic animals. In this species development of the flagellates in the insect is restricted to the proboscis and cibarium where the parasite passes through 2 forms including both the epimastigote and trypomastigote stage. The entire life cycle of this species in the tsetse fly-vector is completed in as little as 3 days after initial infection. Forms of T. vivax in the blood stream of mammals are only of the monomorphic trypomastigote type. Representatives of this subgenus have evolved independently of tsetse flies and have adapted to mechanical transmission as noted earlier.
2) Trypanosoma congolense group (Nannomonas subgenus). Species included in this subgenus use a larger area of the digestive tract of the tsetse fly relative to species in the Dutonnela subgenus: In the gut, the ingested blood trypomastigotes differentiate into long trypomastigotes that migrate to cibarium and the proboscis of the flies where they differentiate into epimastigotes and, then, into metacyclic forms. Like T. vivax, mammalian blood stream trypomastigotes are also monomorphic.
3) Except for T. evansi and T. equiperdum, the representative of T. brucei group (Trypanozoon subgenus) are pleomorphic and go through a much more complex cycle in the vector. There are 2 proliferative forms in the fly, procyclic trypomastigotes in the gut and epimastigotes in the salivary gland and the entire life cycle can be completed in as little as 3 weeks (Sharma et al., 2009).

Figure 3. A female tsetse fly (Glossina morsitans morsitans) from the Serap Aksoy Lab colony at Yale University School of Public Health. Tsteste flies transmit African trypanosomiasis. (Note: Taken by Geoffrey M. Attardo, this photo was a winner of the 2010 Fogarty Grantee Photo Contest, Fogarty International Center, United States, National Institutes of Health.)
(Source: G. M. Attardo, 2010. License: CC BY-NC-SA 4.0.)
Salivarian trypanosomes may be highly pathogenic and lethal for their mammalian hosts, but a wide range of host species are tolerant to several of these trypanosome species. This is the case of native cattle breeds N’Dama and the West African shorthorn (WASH) (Ganyo et al., 2018). The basis of this tolerance, although much studied with several hypotheses already formulated, is still a controversial subject. One unquestionable point is that whether the animals are trypanotolerant and trypanosusceptible is defined by their capacity to control anemia, which is the major outcome of the infection, and that determines whether or not host-animals remain competitive and productive.
The salivarian trypanosomes are not able to invade cells, and only extracellular trypomastigote and epimastigote forms are recognized. Trypomastigote forms are maintained in blood and body fluids of the infected mammal host. In the trypomastigote forms, 3 characteristics can be highlighted:
1) Two main types of blood trypomastigotes can be observed in most of the parasites from this section, including: a) Thin or slender forms are the dividing stage that are completely adapted to the mammal host, and b) broad or stumpy forms do not divide and possess well-developed mitochondria throughout the cell, and are adapted to the cycle in the vector dipterous host.
2) In the mammal host, where access to glucose is unrestricted, the mitochondrion is reduced in size and complexity and the parasite performs compartmentalized glycolysis, that is, within glycosomes. In this organelle, glucose is broken down into pyruvate and ATP, which is very effective whereas in the vector, where glucose is much more scarce, the now active mitochondria carries out an oxidative catabolism cycle generating CO2, H2O, and ATP oxidative cycle that results in CO2 and H2O; which is a more efficient mode of metabolizing glucose. Recently, it was shown that blood forms of Trypanosoma brucei are able to perform gluconeogenesis using glycerol as substrate for ATP production. The implications of this metabolic pathway are still unknown, but it demonstrates an important physiological plasticity. This pathway was suggested as being related to the passage of the parasite through tissues of its multiple host species (Kovařová et al., 2018). In the same way, gluconeogenesis was proposed as an important ATP source and is used by the procyclic forms in the midgut of the tsetse fly vector. In this environment, the parasite uses proline as substrate (Wargnies et al., 2018).
3) The ability of trypanosomes to sequentially modify their surface glycoproteins is an efficient escape mechanism from the mammalian host immune system. This variation in the surface coat of the protozoan are known as VSGs (Variable Surface Glycoproteins) initially described as VATs (Variable Antigen Types) (Barry et al., 1979). Salivarian trypanosomes live an extracellular existence in the blood and lymphatic system of the mammalian host and, are therefore, completely exposed to the humoral immune response, these parasites evolved to periodically alter all of their surface glycoproteins, as if changing a coat, in an efficient programmed mechanism where only one VSG is expressed at a time and is never repeated. It is estimated that there are about 10 million molecules anchored on the surface of these trypanosomatids being periodically exchanged, and about 2,000 vsg encoding genes regulating this process (Mugnier et al., 2016; Romero-Meza and Mugnier, 2020). This phenomenon is responsible for the parasitemia waves characteristic of mammals infected by salivarian trypanosomes. Also, their high motility, capacity of supporting non-immune defense mechanisms, and the mechanical forces inherent to blood circulation are important to survival in the mammal host (Stijlemans et al., 2016). The metacyclic forms of T. brucei develop in the salivary glands of the tsetse flies and also synthesize VSGs that; however, differ from the VSGs from the blood trypomastigotes (Kolev et al., 2017; Romero-Meza and Mugnier, 2020).
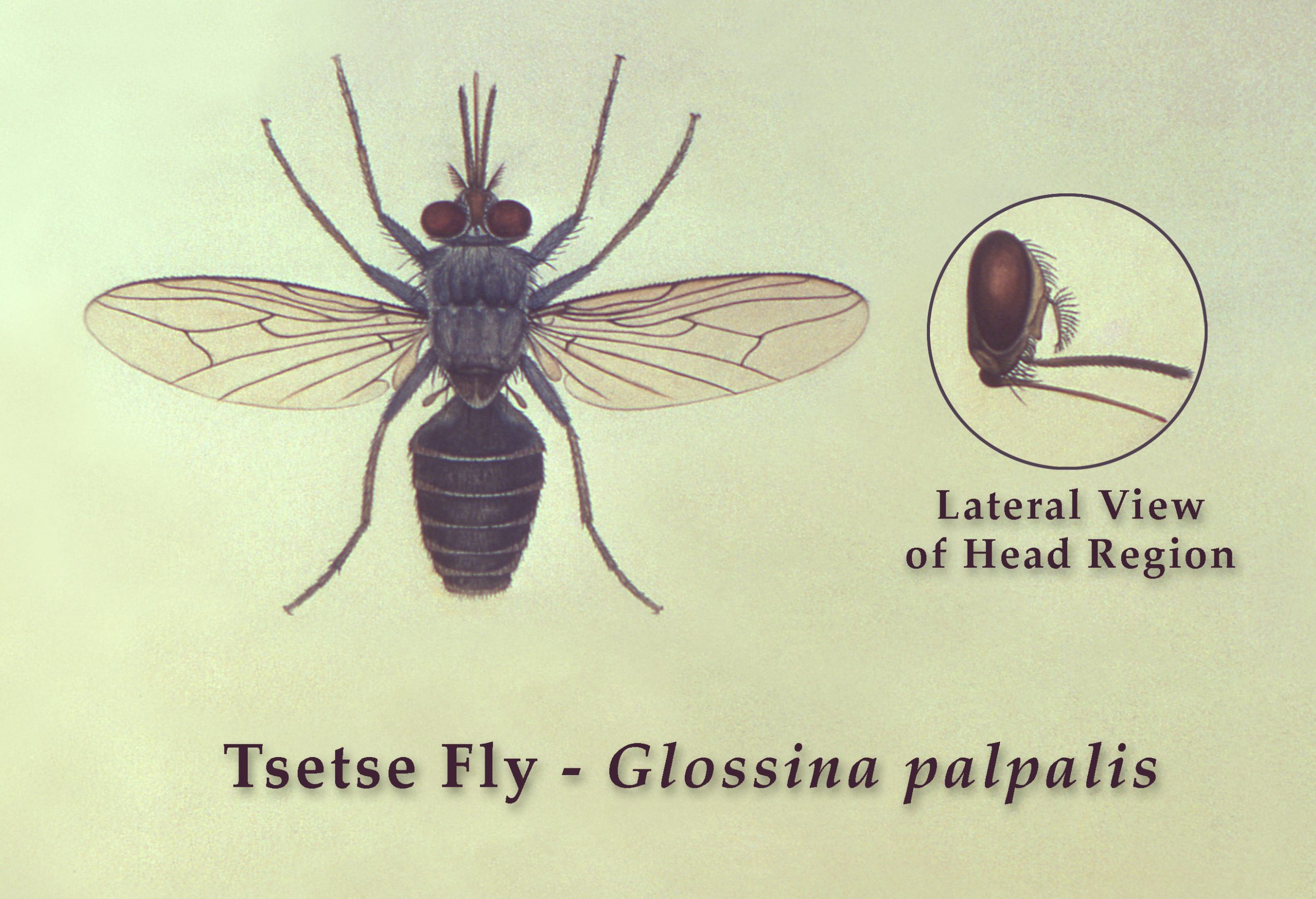
Figure 4. Tsetse fly, Glossina palpalis, top view with lateral view of head region.
(Source: United States Public Health Image library, image 17638; R. Darsie, 1976. Public domain.)
Tsetse flies live in moist savannah and woodlands, regions of more than 500 mm of rain a year, which include more than 30 countries across Africa (Cecchi et al., 2015). In those areas, human sleeping sickness is well-known and is caused by Trypanosoma brucei rhodensiense while T. b. gambiense causes Nagana, the non-human disease that is generally fatal in livestock and cycles in wild mammals (Büscher et al., 2017).
Humans are protected from infection by most of the African trypanosomes because of their production of a trypanolytic protein named Apolipoprotein 1 (APOL1), which is secreted by 2 protein complexes (TLF-1 and TLF-2) leading the formation of pores in the parasite membrane, resulting in its lysis. However, the 2 subspecies of Trypanosoma brucei associated with human sleeping sickness produce substances that are capable of lysing APOL1, the RAS factor in T. b. rhodesiense and TgsGP in T. b. gambiense (Capewell et al., 2015). Interestingly, baboons and one African human population named G1/G2 present a mutation in APOL1 that confers resistance to RAS and, therefore, to the infection by T. b. rhodesiense. The rare cases of humans that become infected with T. b. brucei and other species of trypanosomes are usually associated with people with some mutations that result in the absence of APOL1 production.
The genetic exchange that occurs solely in the insect vector of the salivarian trypanosomes, besides their huge repertoire of surface antigens, implies that new genotypes of salivarian trypanosomes may emerge to infect humans, and domestic and wild animals, and pose an important and worldwide health risk that would be magnified due to the absence of vaccines (Gibson, 2015).
Trypanosoma (Nanomonnas) congolense
Nannomonas trypanosomes include species that infect wild and domestic suidae (Trypanosoma simiae and T. godfreyi), and T. congolense, a parasite that infects a broad spectrum of domestic and wild mammalian species and is the main cause of Nagana in Africa (Hamill et al., 2013; Morrison et al., 2016). These parasites are restricted to areas of occurrence of tsetse flies in sub-Saharan Africa and are described as extremely pathogenic for mammals (Cecchi et al., 2015). Some African breeds (Djallonke sheep, N’Dama cattle, and West African dwarf goats) are trypanotolerant, meaning that they are able to support the infection without anti-therapy and still maintain good health. The mechanisms underlying this tolerance probably depend on a genetic basis but this is still under debate (Yaro et al., 2016).
The trypomastigote forms are characterized by a flagellum that runs through the body of the parasite with a very short free end. The infection often starts with a skin lesion, a canker, where the parasites multiply, before reaching the bloodstream and lymphatic vessels of the host. In the vector, the multiplication takes place throughout the digestive tract before migration to the salivary gland, where the differentiation to metacyclic forms occurs (Dyer et al., 2013).
Human infection by Trypanosoma congolense has been only rarely reported (Truc, 1996). Compared to other African trypanosomes that infect livestock, T. congolense is considered as the most pathogenic, most prevalent, and most widely distributed trypanosomatid within the area of occurrence of Glossina species. Three different genotypes of T. congolense are recognized: 1) The genotype Savannah, which is the most pathogenic and can affect a greater diversity of hosts, including carnivores; 2) the genotype Forest, described as poorly pathogenic and more often observed in cattle, goats, pigs, and dogs; and 3) the genotype Kilifi, considered as non-pathogenic and found infecting domestic ruminants (Rodrigues et al., 2014).
Trypanosoma (Trypanozoon) brucei
Trypanosoma brucei is the etiological agent of sleeping sickness in Africa and one of the few parasites able to cross the human blood-brain barrier, resulting in the nervous symptomatology observed in human disease. This parasite displays 3 subspecies: 1) T. b. gambiense, present in West Africa, associated with the more chronic form of human sleeping sickness, reported as less pathogenic; 2) T. b. rhodensiense, present in East Africa, associated with the acute form, the most pathogenic form of human disease; and 3) T. b. brucei, which infects wild and livestock animals, is associated with Nagana and is only rarely associated with human disease (Büscher et al., 2017).
The initial and recurrent symptoms of sleeping sickness are fever, tremors, muscle and joint pain, lymphadenopathy, malaise, weight loss, anemia, and thrombocytopenia. Later, neurological symptoms and meningoencephalitis can be present associated with mental retardation, convulsions, somnolence, and apathy that can progress to coma and death. This serious human disease is endemic in 36 African countries, including some epidemic areas in Angola, Democratic Republic of the Congo, and Sudan, and affects mainly people living in rural areas where tsetse fly is present. About 95% of human infections are caused by Trypanosoma brucei gambiense and commonly associated with Glossina palpalis. Those cases have a slower evolution of the disease and the infection can be present by months and even years without clinical signs. However, when the first signs appear, the disease is usually already in an advanced state and with the central nervous system compromised. The other 5% of the infections are caused by the T. b. rhodensiense and usually transmitted by G. morsitans. In these latter cases, the infection course is much faster and in a few months or even in a few weeks the disease progresses to the central nervous system presenting the characteristic neurological symptoms (Büscher et al., 2017).
In mammals, Trypanosoma brucei is always present in the extracellular trypomastigote forms and 2 morphotypes are recognized: 1) The slender forms, the divisionary forms that perform glycolysis in glycosomes and cannot survive in the insect vector; and 2) the stumpy forms, that do not divide in blood vessels, and display well developed mitochondrial ridges that are essential for survival in the vector.
Inside vectors, Trypanosoma brucei can colonize the whole digestive tract where both procyclic trypomastigotes and epimastigotes are present and can replicate themselves. The epimastigote forms that reach the salivary glands adhere to the microvilli of glands and perform cell division with subsequent differentiation to pre-metacyclic trypomastigotes, still attached to the salivary gland, but with fewer adhesion plaques. These pre-metacyclic trypomastigotes start to reacquire the coat of glycoproteins that are important in the blood phase of the infection. This form, named nascent metacyclic form, begins to detach itself from the glandular epithelium and will be totally free in the salivary gland when this differentiation is complete. At this point, the glycosomes are reverted to their spherical shape and the mitochondria are reduced to a small structure, which will no longer be functional while this parasite is in its mammalian host. The transmission will take place through the inoculation of these metacyclic trypomastigotes in the following blood meal (Vickerman et al., 1988; Dyer et al., 2013).
Trypanosoma (Dutonella) vivax
Trypanosoma vivax has a wide diversity of ungulate hosts; especially ruminants (order Artiodactyla). The infection occurs in Africa, Asia, Central America, and South America. Both wild and domestic ungulates (including buffaloes and antelopes); as well as equines and camels, are their most common hosts. Infection in laboratory animals has never been established (Osório et al., 2008). This parasite species can be transmitted both in cyclic and mechanical transmission. Cyclic transmission is reported in Africa, in areas where tsetse flies are present, and usually results in a more severe form of the disease. Mechanical transmission occurs in other parts of Africa (apart from the tsetse geographical area), in addition to Asia and the Americas, where the disease tends to be milder in cattle. In Brazil for example, outbreaks have always been associated with low mortality and few economic losses (Silva et al., 1996; Batista et al., 2007; Bastos et al., 2017).
In the mammalian host the trypomastigote forms are found exclusively in the blood. In Glossina species, where the transmission is cyclic, this parasite differs in the replicative epimastigote forms that are restricted to earlier parts of the digestive tract. After the differentiation to trypomastigote forms, parasites migrate from the hypopharynx to the salivary gland where they differentiate into metacyclic trypomastigotes, which are the infective forms inoculated by the tsetse. Once in the salivary gland, Trypanosoma vivax can be maintained throughout the whole life of the vector. In mechanical transmission, there is no differentiation and the trypomastigote forms are carried from one mammal host to another by other hematophagous flies, especially those from the Stomoxys and Tabanus genera.
The first symptoms of Trypanosoma vivax infection are usually unspecific, such as anemia, fever, apathy, weight loss, and diarrhea. Diverse reproductive problems are associated with the infection, including transplacental transmission and abortion. When present, the neurological symptoms observed are incoordination, muscle tremors, transient and/or permanent
blindness, meningoencephalitis, and malacia (Batista et
al., 2007; 2009).
Trypanosoma vivax was introduced into the Americas, probably with cattle brought from the European colonies in Africa. On this new continent, the parasite adapted to the mechanical transmission by several hematophagous insects, circulating among recently introduced domestic cattle and, perhaps, has spread among wild ungulates such as the cervids, never before exposed to this parasite. In Brazil, the first report of the parasite was in a buffalo in the swampy regions of Marajó Island, in the Brazilian Amazon (Shaw and Lainson, 1972). Later, in the late 1970s, this parasite was reported in sheep and cattle in the State of Amapá, also in Amazon region, and only a decade after this reported outside the northern region of the country, in cattle from the Brazilian Pantanal biome (Silva et al., 1996).
Possibly, the first outbreaks in the Pantanal were due to the increase of the displacement of animals from the north to the center-west of Brazil. In this biome, Trypanosoma vivax epidemiology is directly associated with drought and flood periods. In flooding periods, the reduction of pasture area increases the animal density per area, resulting in nutritional problems and resulting in higher susceptibility to infections, including trypanosomiasis (Silva et al., 1996).
Trypanosoma vivax can also be pathogenic for horses (da Silva et al., 2011). In both natural and experimental conditions, asinins were demonstrated to present high infection rates in subpatent and asymptomatic infections (Rodrigues et al., 2015). The infection of domestic ruminants by T. vivax results in severe economic losses, especially in South America. In spite of this, T. vivax remains poorly studied as a consequence of the inability to grow this trypanosome species in mice or in culture media.
Trypanosoma (Trypanozoon) evansi
The species Trypanosoma evansi masterfully exemplifies the genetic plasticity and consequent evolutionary success of salivarian trypanosomes. Together with T. equiperdum, T. evansi seems to have branched off from T. brucei due to the profound alterations of its kinetoplast DNA and, as a consequence, to have gained independence from a biological vector and to be transmitted mechanically, which has resulted in its enormous dispersion throughout Asia, the Americas, and Africa. The distinct kDNA alteration patterns observed in T. evansi samples suggest that T. evansi arose multiple times from a different T. brucei ancestor. These findings point to the necessity of revisiting the nomenclature of the members of the subgenus Trypanozoon (Radwanska et al., 2018).
Among all trypanosomes, Trypanosoma evansi is the one that is able to infect the largest variety of mammalian hosts, being dispersed in all continents (Desquesnes et al., 2013a). This parasite is the etiological agent of one of the major diseases affecting horses, called Surra (in the Old World) or Mal de Caderas, Quebra Bunda, or Derrengadera (in South America). The transmission is mechanical, being carried out normally by hematophagous flies from Tabanus species and Stomoxys species (Desquesnes et al., 2013b). The T. evansi infection has been recently considered by OIE (World Organization for Animal Health) as a mandatory disease (Jaimes-Dueñez et al. 2017). Human cases are rare, but have been reported in Africa and Asia, usually associated with an extremely rare condition termed Tangier disease (Tomlinson et al., 1995).
Trypanosoma evansi belongs to the same Trypanozoon subgenus of the T. brucei species but is different from T. brucei whose transmission is totally dependent on cyclic transmission. Trypanosoma evansi is dependent only on mechanical transmission, even by tsetse flies. That is because, at some point on the evolutionary path of these parasites, T. evansi differentiated from T. brucei and lost the kDNA maxi-circles where most of the genes responsible for the oxidative metabolism and multiplication are located in Glossina species, making T. evansi incapable of multiplying in the biological vector. Currently, some researchers suggest that this species comprises total or partial diskynetoplastic T. brucei strains. Although still considered to be a different species, some authors propose that T. evansi (and T. equiperdum) be classified as subspecies of T. brucei, or even variant strains of T. b. brucei (Carnes et al. 2015; Wen et al. 2016).
Trypanosoma evansi is a monomorphic parasite, and the trypomastigote form is the only recognized morphotype. Its trypomastigotes are the replicative forms observed exclusively in the blood of infected mammals and its morphology is exactly identical to the stumpy forms of T. brucei. Infection in mammals usually results in very high parasitemias, favoring mechanical transmission. Besides the mechanical transmission by hematophagous flies and the iatrogenic form through sharing of contaminated fomites, hematophagous bats display a differentiated feature, being able to act both as reservoir and vector of this parasite. That is because, in addition to being infected as all other mammalian hosts (which means that the parasite multiplies in the blood of these animals), it can still transmit the parasite that is easily present in its saliva during a blood meal (Desquesnes et al., 2013b). The oral route and agonistic encounters are also proposed as transmission routes in the wild (Herrera et al., 2011).
The estimated date of entry into South America by Trypanosoma evansi is still controversial. Although it is hypothesized that T. evansi was introduced by infected horses of European colonizers (Desquesnes et al., 2013a), other authors propose that their introduction was much earlier, perhaps carried by the first primates and caviomorph rodents that came directly from Africa to South America, about 35–40 Ma (= million years ago). Caviomorph rodents, including capybaras, are considered to be important T. evansi reservoirs in the Pantanal region (Herrera et al., 2004) and infected rodents may have entered the Americas through the well described island-hopping or even sweepstakes dispersal processes (Lavocat, 1974; Raven and Axelrod, 1975; Flynn and Wyss, 1998).
In South America, trypanosomiasis caused by Trypanosoma evansi is economically very important in the flooded areas of the Brazilian Pantanal and Argentinean Chaco regions. These regions have a great concentration of livestock, and horses are essential for cattle handling. Outbreaks occur sporadically, though more common after a flood period, and result in a diversity of clinical features, from mild to severe fulminant forms (Herrera et al., 2004). In most severe cases, horses may present with anemia, emaciation, and subcutaneous edema of the lower body regions, with reports of abortion in pregnant females, and various types of neurological manifestations. The characteristic symptoms that inspired the name of the horse disease are the atrophy of the great muscular masses of the pelvic limbs, devolving to incoordination and ataxia (Desquesnes et al., 2013a).
The main wild reservoirs in the Pantanal region are the coatis and capybaras because they: 1) Are quite abundant in the region; 2) present high infection rates with long-lasting patent parasitemia; and 3) may remain infected for a long time. Capybaras support infection by T. evansi without anemia and while maintaining good general health. Coatis, in contrast, when infected by T. evansi display anemia (Herrera et al., 2002; 2004). Infected horses may also act as reservoirs because the persistence of the parasite in asymptomatic animals after treatment is not rare (Herrera et al., 2004).
Usually reported in domestic animals around the world, studies on Trypanosoma evansi transmission in the wild are generally restricted to the Brazilian Pantanal. In this region, T. evansi can be considered a typical enzooty, with several infected wild mammals already found and occurring in sympatry with other trypanosomes, such as T. cruzi and T. rangeli. Interestingly, most infections observed in the wild seem to be subpatent and anemia is not commonly observed (Rademaker et al., 2009).
Trypanosoma (Trypanozoon) equiperdum
The other species within the Trypanozoon subgenus, also considered as a subspecies of Trypanosoma brucei or a mutant strain of Trypanosoma b. brucei by some authors (Carnes et al., 2015; Wen et al., 2016), is Trypanosoma equiperdum. This species is the causative agent of equine Dourine, a disease that affects horses and other equidae. The transmission is exclusively venereal and the parasite is found only in genitals and their secretions. Asinines are usually reported as asymptomatic carriers of the parasite (Gizaw et al., 2017). Although as widespread as Trypanosoma evansi in the world, the reports of infection are quite intermittent and most studies are only case reports. The symptoms are similar to other trypanosomiases, such as fever, anemia, and emaciation, besides symptoms more specific to the genital organs, such as edema of the genitalia and mammary glands. More severe forms may progress to incoordination, facial paralysis, and death (Gizaw et al., 2017).
The difficulty in studying Trypanosoma equiperdum is that there are almost no available isolates of this parasite, and most isolates are from the beginning of the last century and lack essential information such as isolation site, year, and even host. After a long time without description of new isolates, in 2015, a group from Venezuela obtained the first 2 isolates of this parasite in Latin America (Sánchez et al., 2015). In 2016, another group obtained a new isolate from the urogenital tract of a horse in Mongolia (Suganuma et al., 2016). The lack of knowledge about T. equiperdum reinforces the question about the validity of this species (or subspecies).
Trypanosoma (Tejeraia) rangeli
One trypanosome that can be classified as neither Stercoraria nor as Salivaria, is Trypanosoma rangeli (see Grisard, 2002). Actually, T. rangeli is a parasite species transmitted by the saliva of its insect vector but shares numerous characteristics with other species of the Stercoraria section, as it is easily cultivated in axenic media (which is not observed in the Salivarian trypanosomes). It also shares mammalian hosts and vectors with T. cruzi. Currently, this parasite species is classified in the subgenus Tejeraia that was created in 1982 only to classify T. rangeli, until then classified within the Herpetosoma subgenus, within the Stercoraria section (Añez, 1982).
Trypanosoma rangeli is a multi-host (mammal) parasite found exclusively in the Americas and is capable of infecting humans. The main vector species are triatomines from the Rhodnius genus, in which T. rangeli differentiates to the infective metacyclic forms in the salivary gland of the insect (Guhl and Vallejo, 2003). Moreover, other triatomine species may also maintain T. rangeli, as in the case of Triatoma vitticeps (see Dario et al., 2017a). The genetic diversity within T. rangeli was first observed based on the difference of a nucleotide sequence from its mini-circles, resulting in the recognition of 2 separate populations, KP1(+) and KP1(−) (Vallejo et al., 2002). Further studies have shown that this genetic polymorphism is even more extensive and 5 lineages could be described, from A to E (Maia da Silva et al., 2009). It was first proposed that the divergence of these lineages was related to different Rhodnius species involved in the transmission, but it is now accepted that this separation of lineages is not strict. In fact, the characterization in distinct lineages is recent and we do not have enough sampling to propose associations between T. rangeli lineages and mammal hosts and/or ecotypes (Urrea et al., 2011; Dario et al., 2017a).
In the mammal host, only the blood trypomastigote form is observed and the current consensus in the scientific community is that Trypanosoma rangeli does not differentiate in amastigote forms, as occurs in T. cruzi. In the Rhodnius vector, the predominant and replicative form is the epimastigote that colonizes the vector’s gut. Some of these parasites differentiate into trypomastigotes in the final portion of the intestine and are eliminated with the feces, but these forms are not infective for mammals. Most of the epimastigote forms invade the vector’s hemocoel, where they can multiply both inside and outside hemocytes. From the hemocoel, some of these parasites reach the salivary gland and differentiate into metacyclic trypomastigotes that are transmitted through the saliva during a blood meal (Guhl and Vallejo, 2003).
Some points of this transmission cycle still need to be clarified. First, only the blood trypomastigote form is described in mammal hosts, which is considered to be non-replicative. Otherwise, this parasite is possibly able to multiply in the mammalian host because there are numerous cases of persistence of parasitism even after long periods after exposure. For instance, Trypanosoma rangeli was isolated from Brazilian patients who had been out of risk areas for many years and were being treated as having Chagas disease (de Sousa et al., 2008). These cases suggest that this parasite species can multiply in the mammal host, in a mechanism still not identified.
Another intriguing aspect is the niche occupied by T. rangeli in mammal hosts. This parasite is always diagnosed in blood, but it has been isolated from bone marrow from an anteater in the Amazon region. The presence of T. rangeli in this unorthodox niche probably occurred through blood or lymph circulation, as described for other trypanosomes, and could had been influenced by the coinfection of T. cruzi and Leishmania infantum that were also diagnosed in the same host (De Araújo et al., 2013).
The Trypanosoma rangeli infection in mammals are considered innocuous or non-pathogenic, although due to the unknown mechanism of parasite multiplication in mammals, the discovery of T. rangeli in bone marrow and the isolation in Chagas disease patients are aspects that have to be considered in the endorsement of this assumption. On the other hand, the pathogenicity of T. rangeli infection in invertebrate hosts is well described and reports of the influence of the parasite load on insect molt and destruction of intestinal epithelium during parasite invasion to the hemocoel are some of the damaging effects usually observed in infected bugs (Ferreira et al., 2010; García et al., 2012).
Section Stercoraria
Trypanosomes of this section include numerous species of Trypanosoma that live in the intercellular spaces and/or in the blood of their mammalian hosts. Most of these do not display mechanisms of colonizing the intracellular environment or disguising immune response by using variable surface antigenic variation as observed in some salivarian trypanosomes. This is the case for several species of Trypanosoma from this section. Stercorarians include heteroxenous parasites that are transmitted between species belonging to all vertebrate classes and hematophagous invertebrates on all continents. Three main evolutionary stages are recognized in this section, including: 1) Extracellular trypomastigote forms, 2) extracellular epimastigote forms, and 3) intracellular amastigote forms. Interestingly, in the stercorarian trypanosomes, replication by cellular division/binary fission of these trypanosomes in mammals occurs only in either the amastigote or the epimastigote stages; fission of the trypomastigote forms in this group is unknown (Hoare, 1972).
All species from this section possess trypomastigote forms with flagella protruding anteriad of the cell with a large and non-terminal kinetoplast. In the vector insects, the final development of the parasites, which corresponds to the formation of metacyclic trypomastigotes, occurs in the posterior portion of the digestive tract of the insect vector host. The main characteristic, which gives the name to the section (stercoraria or posterior station), is their transmission method which is always via insects’ feces by rubbing metacyclic trypomastigotes into a wound, eye mucus membranes, or oral mucus membranes (Hoare, 1972).
Trypanosoma (Herpetosoma) lewisi
The subgenus Herpetosoma comprises all the species described in the Trypanosoma lewisi group, which is the type species of the subgenus. Species from this subgenus include trypanosomes from many species of rodents, in addition to a lagomorph trypanosome called T. nabiasi. Most known vectors are fleas, but for many Trypanosoma (Herpetosoma) species, the transmission cycle is still completely unknown.
Trypanosoma lewisi is a cosmopolitan parasite of Rattus species. This trypanosomatid species was probably introduced to the different continents and countries due to its association with Rattus rattus, a synanthropic rodent that in turn has accompanied humans since their first voyages. Trypanosoma lewisi was previously considered specific to Rattus species. and not capable of infecting the other cosmopolitan rodents (Mus musculus) in experimental conditions. Currently, it is recorded in some wild rodent species (Mafie et al., 2019), besides primates, including humans (de Sousa, 2014). The phylogenetic analysis of T. lewisi isolates from Rattus species. and primates proposed that T. lewisi underwent a process of host switching between rodents and primates through accidental contamination of primates with infected fleas from these rodents (Maia da Silva et al., 2010). Some cases have been reported in Asia, associated with fever, anemia, and immunosuppression (Verma et al., 2011).
Some authors consider T. lewisi a neglected re-emerging human pathogen (Lin et al., 2015) in spite of the rarity of cases of human infection by T. lewisi worldwide and because it always presents in immune incompetent individuals or people living in close contact with rats (Verma et al., 2011; Shah et al., 2011). The recognized vectors of Trypanosoma lewisi are Xenopsylla cheopis and Nosopsyllus species, but it is believed that other fleas, such as Ctenocephalides canis, Leptopsylla segnis, and Pulex irritans can also act as vectors (Hoare, 1972). No cell invasion is reported in vertebrates, and the infection is considered innocuous in immunocompetent hosts, although it can be lethal in newborn rats or increase susceptibility to other parasites when presenting as a co-infection, as with Toxoplasma gondii (Ríos Carrera et al., 2009) or Cryptococcus neoformans (Gross et al., 2006).
Two stages of the parasite can be observed in a vertebrate’s blood, the replicative epimastigote forms, and the trypomastigote forms. Inside the digestive tract of fleas, the trypomastigote forms invade the epithelial cells of the stomach and differentiate into amastigote forms, which are replicative, and differentiate again to trypomastigote forms before returning to the digestive lumen. After reaching the flea’s midgut, the trypomastigote forms differentiate into replicative epimastigotes that will differentiate into metacyclic trypomastigotes at the end of the digestive tract. Besides the contaminative route, oral transmission through accidental ingestion of fleas has been demonstrated for Trypanosoma lewisi and other species from the same subgenus, namely, T. microti, T. evotomys, and T. grosi (see Maraghi et al., 1995).
In mammals, infection by Trypanosoma (Herpetosoma) species is characterized by intense and short parasitemias. The infection is easily controlled by the host in about 3 weeks due to a humoral immune response because the parasite has limited capacity for antigenic variation and does not invade mammal cells. A characteristic observed in T. lewisi, and also believed to occur in other species from the same subgenus, is the production of an IgG immunoglobulin named ablastin that inactivates the replicative forms of the parasite leading to the abrupt remission of the parasitemia. After this, rodents become resistant to new infections (Drew and Jenkin, 1982). The abrupt remission of parasitemia occurs only when preceded by an initial phase of infection establishment where, most probably, other factors are involved. The passive transfer of ablastin from one infected rodent to another was observed, which resulted in a partial control of the infection, but not as abruptly as observed in natural infections (Drew and Jenkin, 1982).
An intriguing aspect of the transmission cycle has been observed in Trypanosoma musculi, a parasite considered restricted to Mus musculus. After the phenomenon of ablastin, rodents infected by T. musculi become resistant to new infections and parasitemia is no longer observed, but throughout their life, the infected rodents still maintain some parasite forms, including replicative forms, in the vasa recta of the kidneys. These forms are biochemically and molecularly different from blood forms and appear to represent a new evolutionary stage of the parasite that are not inactivated by the host’s immune system, due to the high concentration of urea (Monroy and Dusanic, 2000). The consequences of the existence of this distinct stage in the life cycle, the occurrence of this phenomenon in other Herpetosoma species, and the evolutionary impact of this parasite persistence are still unknown aspects.
Besides Trypanosoma lewisi and T. musculi, some other related species described in the same Herpetosoma subgenus are: T. microti, described in Microtus spp. and also directly transmitted through agonistic contact in the reproductive season; T. evotomys and T. grosi from Old World rodents; T. kuseli and T. otospermophili from squirrels; and T. nabiasi, a parasite species specific of the lagomorph Oryctolagus cuniculus present in the New World and Australia, the latter as a result of the human introduction of infected fleas in an attempt to control the rabbits (introduced one century before) with the Myxomatosis virus (Hamilton et al., 2005).
Trypanosoma (Megatrypanum) theileri
The morphology of parasites from the subgenus Megatrypanum is very typical: they have very large cells with a visible undulating flagellum that adheres to the entire body of the parasite. Trypanosomes from this subgenus are transmitted by a diversity of vectors, including hematophagous flies, fleas, ticks, and Pseudolynchia species. This subgenus comprises species that infect a wide variety of hosts, including marsupials, rodents, primates, and, mainly, varieties of bovinae cattle (Kingston, 1991; Kelly et al., 2017). The species Trypanosoma (Megatrypanum) theileri is a hemoparasite found in Artiodactyla species worldwide and is the type species of the subgenus Megatrypanum. Recently, a second escape mechanism of modeling the trypanosome cell surface, distinct from the VSG’s coat of the salivarian trypanosomes, has been described. Actually, the surface of Trypanosoma theileri cells can be modeled by the expression of a mixture of proteins (Kelly et al., 2017).
The transmission cycle of Trypanosoma theileri is not completely elucidated yet. Mammals are proposed to have a replicative epimastigote form, but this parasite form is very rarely observed in field studies, probably because these forms quickly differentiate into trypomastigotes, which is the predominant morphological type detected in mammals’ blood. In mammals, those trypomastigote forms are capable of invading all body fluids, in addition to some tissues, such as lymph nodes, kidney, spleen, and brain. For long time, it was accepted that T. theileri did not differentiate into amastigotes, but the existence of these stage forms was recently demonstrated in vitro, including the ability to differentiate and invade other cells in a process similar to that observed in T. cruzi (see Lee et al., 2013). In vectors, the epimastigote forms of the parasite replicate in the gut before differentiating into metacyclic trypomastigotes that are eliminated in the feces. The tabanids are the vectors of T. theileri, but a tick species (Hyalomma anatolicum) was proposed before as an alternative vector (Latif et al., 2004). As observed for other Stercorarian trypanosomes, oral transmission through accidental ingestion of infected vectors is proposed as an important infection route (Kelly et al., 2017).
Trypanosoma theileri is a cosmopolitan and opportunistic parasite. The parasitemia in bovines is usually associated with some unspecific clinical signs. Infection may remain subpatent and asymptomatic for several years but may result in high parasitaemia and pathogenicity when associated with other factors, such as stress, malnutrition, or pregnancy. This parasite species is recognized in bovines and buffaloes from South America, and at least 10 genotypes are recognized, most of them specific in terms of mammal species (García et al., 2011).
Besides Trypanosoma theileri, some other species from the same Megatrypanum subgenus are: T. minasense from New World primates; T. melophagium from sheep; T. theodori from goats; T. cervi and T. mazamarum from cervids; T. freitasi that colonize scent glands of Didelphis species; T. samueli and T. saloboense, from Monodelphis sp. from the Amazon region; T. peba from the 6-banded armadillo Euphractus sexcinctus; T. legeri from the anteater Tamandua tetradactyla; Trypanosoma pestanai isolated from European badgers; and Tr. caimani from alligators from the Brazilian Pantanal; as well as a high diversity of bird trypanosomes. For the less studied Megatrypanum species, other vectors (apart from tabanids) are involved in the transmission: keds (Melophagus ovinus) are involved in Tr. melophagium transmission (Gibson et al., 2010), Tr. pestanai is transmitted by fleas (Peirce and Neal, 1974; Lizundia et al., 2011), while Tr. cervi are also spread by keds (Böse and Petersen, 1991). It is worth mentioning that many of these parasite species were described based on morphology, before the employment of molecular techniques for parasite identification. As a result, some species may not be valid, as in the case of Tr. saimirii and Tr. leeuwenhoeki, currently recognized as synonyms of Tr. rangeli (See Stevens et al., 1999a; Ziccardi et al., 2005).
Trypanosoma (Schizotrypanum) Species and Other Species Related to T. cruzi
Almost all the Trypanosoma (Schizotrypanum) species are parasites exclusive of bats. The exceptions are T. cruzi, a multihost parasite that may also infect bats, and T. dionisii, a parasite species considered specific to bats, but recently observed also in human cardiac tissue and in the marsupial Monodelphis americana (See Lima et al., 2015a; 2015b; Dario et al., 2016; 2017a; 2017b). Parasites from this subgenus are morphologically identical and all of them are able to differentiate into metacyclic forms and invade cells, where they differentiate into amastigotes.
Until the 1990s, most Trypanosoma infections in bats were described as T. cruzi-like because of the difficulties differentiating among Schizotrypanum species. Currently, due to advances of molecular techniques, especially since it is easier now to analyze DNA sequences, the variety of Trypanosoma species infecting bats has been shown to be enormous, and this diversity has been increasing annually with descriptions of new Trypanosoma species infecting bats. Some Trypanosoma species reported in bats are: T. cruzi marinkellei and T. wauwau, described in bats from Central America and South America; T. vesperitilionis and T. dionisii, described in bats from both the Old World and the New World; T. pipistrelli from Old World bats; T. pteropi, T. hipposideri, and T. teixeirae from Australian bats, including the enormous flying foxes; T. hedricki and T. myoti from bats surveyed in North America; T. erneyi and T. livingstonei from Africa; and yet others (Lima et al., 2012; 2013; 2015a; 2015b).
The majority of the descriptions of Trypanosoma infection derived from insectivorous bats suggest that, besides the vectorial contaminative route, oral infections must also occur (Lima et al., 2015a; 2015b; Dos Santos et al., 2018). The vectors involved in the majority of bat trypanosome transmission are unknown. Trypanosoma cruzi marinkellei is currently recognized as a subspecies of T. cruzi and reported to be transmitted only by triatomines of the genus Cavernicola, associated with caves (Marinkelle, 1982). Vectors of T. dionisii and T. vespertilionis are reported to be cimicids, but both T. c. marinkellei and T. dionisii were recently found infecting Triatoma vitticeps, a triatomine bug very common in the Atlantic rainforest (Dario et al., 2017a). The close association of Schizotrypanum parasites and bats suggests a long co-evolutionary process. However, the association with different vectors points to an independent evolution between species within this subgenus.
Other Trypanosoma species more recently described were not classified in any subgenus but are phylogenetically closer to T. cruzi and known as trypanosomes of the T. cruzi clade. Besides all the above mentioned trypanosomes of bats, this clade includes T. noyesi, a trypanosome described from the Australian endangered woylie (Bettongia pencillata); 2 isolates from African terrestrial mammals (1 feline and 1 primate); T. conorhini, a species taxonomically classified in the subgenus Megatrypanum, but phylogenetically in the T. cruzi clade; and T. janseni, a species recently described in Brazilian opossums that, along with T. wauwau, forms a sister group of the trypanosomes found in Australian marsupials (Hamilton et al., 2009; Botero et al., 2016; Lopes et al., 2018).
From these, one of the most studied is Trypanosoma conorhini. This species infects Rattus rattus a synanthropic rodent and the kissing bug Triatoma rubrofasciatta all over the world, and only experimentally was demonstrated to be able to infect Mus musculus and Macaca mulatta (Deane et al., 1986). The only parasite form observed in the rodent’s blood is the trypomastigote, without evidence of cell invasion and differentiation to amastigote forms. The biological cycle in the vector includes the replication of the epimastigote forms in the bug’s gut and differentiation in metacyclic trypomastigotes that are eliminated in the feces (Hoare, 1972). The fact that this trio–Rattus rattus, Triatoma rubrofasciatta, and Trypanosoma conorhini–are found together in different parts of the world indicate that their dispersal process occurred together, probably from Asia, which represents a unique joint migration process within the heteroxenous protozoa (Deane et al., 1986).
In general, the vectors involved in the transmission of most of the Trypanosoma species from the T. cruzi clade are unknown, but all of the vectors described up to now are hematophagous hemipterans. In fact, the knowledge of the diversity of Trypanosoma species has been a growing subject of study, especially in the past decade when DNA sequencing tools became more accessible. These studies have shown a much greater diversity than was previously known and that the evolutionary paths within this clade are still not fully understood.
Trypanosoma cruzi
Trypanosoma cruzi is the etiological agent of one of the most important and neglected parasitic disease, Chagas disease (Chagas, 1909). However, the human infection is only a minor trait of this parasite ecology that is a wild animal parasite maintained among dozens of species of mammals and triatomines (family Reduviidae, subfamily Triatomiane). In fact, T. cruzi is a multi-host parasite, able to infect virtually any mammal species and, within them, any nucleated cell (Jansen et al., 2015). In spite of presenting a population structure that is basically clonal, mitochondrial introgression events are not unusual and evidence of genetic exchange has already been described in T. cruzi. Also, recombinant strains have been described (Lewis et al., 2011).
There are 2 non-mutually exclusive hypotheses that explain the origin of Trypanosoma cruzi. The first, called Southern Supercontinent, places the origin of this parasite from an ancestral trypanosome of marsupials, which represented the dominant local fauna, in a supercontinent formed by South America, Antarctica, and Australia. This hypothesis is supported by: 1) The presence of a trypanosome related to T. cruzi, named T. noyesi (Botero et al., 2016), isolated from an Australian woylie (besides other related isolates from Australian marsupials); 2) the estimated divergence between T. cruzi and T. brucei (approximately 100 million years) that is the approximate time of separation of this supercontinent from Africa (Stevens et al., 1999b); and 3) the recent description of T. janseni, which was described in Brazilian opossums and clusters with T. wauwau in a well-supported clade, representing a sister group of the trypanosomes found in Australian marsupials (Lopes et al., 2018).
The second hypothesis points to the origin of Trypanosoma cruzi from ancestral Trypanosoma species of bats and is called the Bat Seeding Hypothesis (Hamilton et al., 2012). This hypothesis is supported mainly by: 1) The increasing evidence of the diversity and polyphyly of bat trypanosomes; and 2) the description of infection of trypanosomes from the T. cruzi clade in African monkeys and palm civets (which does not represent a single lineage that could have been introduced more recently; that is, post-separation of the continents) (Hamilton et al., 2009). Several processes of trypanosome spillover from bats to terrestrial mammals must have occurred until one of them was successfully established and the evolution of this parasite resulted in a new species currently known as T. cruzi (Hamilton et al., 2012).
The contaminative transmission of the parasite, described as the classic form of transmission, occurs when the insect vector eliminates metacyclic trypomastigote forms with feces during a blood meal. These parasites penetrate through lesioned skin or mucous membranes or when the person scratches the location of the insect bite. In the mammal host, these parasites invade the nucleated cells, in which they differentiate into the replicative amastigote forms. A novel differentiation into blood trypomastigotes occurs before the cellular disruption that results in the release of these forms that can invade other cells or circulate in blood vessels and be ingested by other Triatominae in a new blood meal. In invertebrate hosts, Trypanosoma cruzi differentiates into the replicative epimastigote forms and only in the final portion of the insect gut, the parasites differentiate into the metacyclic trypomastigote forms that are the infective forms eliminated in the feces.
This classical route of transmission, however, does not reflect all possible routes of infection in mammals, including humans. From the 4 evolutionary stages of the parasite, only epimastigotes may not be infective, mainly when they are in the exponential growth phase. Moreover, epimastigotes of the stationary phase are already resistant to the complement system and already infective (Kessler et al., 2017). Nutrient impoverishment and starvation of the parasite acts as a trigger for metacyclogenesis (Barisón et al., 2017). Human infections can also occur during blood transfusion and organ transplantation, and these are the most important infection routes in non-endemic countries such as the United States, Spain, and Japan (Gascon et al., 2010). The congenital form, well characterized in humans, seems to have a more regionalized importance, especially in the southern region of South America and non-endemic areas. The oral route has emerged as a very important transmission route in the wild and has been responsible for an increasing number of human infections in the past few decades (Coura, 2015). In the South American Amazonian area, especially in Brazil, this transmission route represents more than 90% of new cases and is usually associated with intake of food contaminated by feces of infected triatomines or infected vectors accidentally crushed along with food, such as sugar cane and açaí juice (Figure 5) PAHO, 2009). This transmission route requires specific surveillance measures whose operation is still not completely known (Xavier et al., 2014).

Figure 5. The oral route has emerged as a very important transmission route in the wild and has been responsible for an increasing number of human infections in the past few decades. In Amazonia, especially Brazil, this transmission route represents more than 90% of new cases and is usually associated with intake of food contaminated by feces of infected triatomines or infected vectors accidentally crushed along with food.
(Source: Pan-American Health Organization, 2009. Permissions: Reproduction is permitted with attribution.)
In nature, the oral route is probably the oldest and most efficient route for parasite transmission and occurs mainly in 2 situations: 1) Ingestion of triatomine feces when the animal scratches the site of an insect bite with its mouth; or 2) predation of infected bugs or mammals. Other possible routes include fights between mammals (when the oral mucosa of a mammal comes in contact with the infected blood of the other mammal) and fomites contaminated by scent gland material of infected Didelphis sp. (Deane et al., 1984; PAHO, 2009), although these are still not empirically confirmed.
This biological plasticity of Trypanosoma cruzi results in a parasite widely dispersed in nature and immersed in transmission cycles that can be characterized as multivariable, complex, and peculiar to each locality (Jansen et al., 2015). Being found in the most diverse ecological niches from the southern parts of Argentina to southern United States, its transmission cycles are quite complex as it includes hundreds of mammalian species and vectors in scenarios of transmission that can be independent and overlapping with each other (Jansen et al., 2018).
As pointed out before, Trypanosoma cruzi has been circulating among wild mammalian fauna for at least 20 million years (depending on which hypothesis for the parasite’s origin is considered). Humans are believed to have existed for no more than 500,000 years. Humans are estimated to have entered into the Americas (and therefore, exposed within this time to T. cruzi transmission cycles) only 15,000 to 24,000 years ago (Bourgeon et al., 2017). As such, there is evidence of human infections in mummies long before the arrival of Europeans in the Americas. Reports in paleoparasitological studies demonstrated the infection in 40% of the mummies examined in Chile, including at least one of them dating to 9,000 years. Almost half of the mummies had signs of megasyndromes (especially megacolon) and infections were seen among mummies from different cultural groups (Aufderheide et al., 2004). It is reported in many South American Aboriginal cultures the habit of feeding on raw or undercooked meat or even drinking the fresh blood of hunted mammals, which would result in infection by T. cruzi (Guhl et al., 2014). The contaminate vectorial transmission probably also occurred: 1) Inside caves used as human dwellings, due to the presence of kissing bugs associated with rocky environments, as in the case of Triatoma brasiliensis (Araújo et al., 2003); and 2) in peridomiciliary environments, as a consequence of the presence of small mammals such as guinea pigs that provided an abundant blood source for bugs in this environment (Aufderheide et al., 2004). These ancient scenarios demonstrate that there are no new or old ways of T. cruzi transmission. Humans have been exposed to the infection by both the contaminative and oral routes since they reached the Americas.
The species Trypanosoma cruzi is a monophyletic and extremely heterogeneous taxon that presents a multiclonal population structure influenced by mechanisms of gene exchange, epigenetic factors, introgression, and others (Stevens et al., 2001; Leonard et al., 2011). The heterogeneity of the parasite has been observed since its discovery, and thin and broad parasite forms observed in patients’ blood were first associated with male and female gametocytes (Chagas, 1909). The first attempts to group T. cruzi isolates with similar characteristics dated from the 1970s: the biodemes, based on a distinct infection pattern in laboratory animals (Andrade et al., 1970) and the zymodemes, based on biochemical patterns of isoenzymes (Miles et al., 1977). The advent of molecular techniques and the proposal of distinct molecular targets in the 1980s and 1990s have revealed that this heterogeneity was much higher than previously thought. In the last decade, however, with the availability of gene sequence analysis in research labs, an even higher diversity of T. cruzi populations (and other related species) has emerged. In this scenario, the most employed gene targets are the small subunit (18S) ribosomal RNA gene and the nuclear glycosomal glyceraldehyde-3-phosphate dehydrogenase (gGAPDH) (Lima et al., 2015a; Lopes et al., 2018). These are the ones with the highest discriminatory power and those targets with the highest number of sequences deposited for comparison. Moreover, more easy accessibility to Next Generation Sequence (NGS) will probably uncover even higher trypanosomatid diversity, especially due the capacity to identify and characterize mixed infections directly on blood samples (Dario et al., 2017b; Pronovost et al., 2020).
Independent of the different molecular targets employed, 2 distinct and phylogenetically distant Trypanosoma cruzi populations have been recognized and were the basis for the first nomenclature consensus for T. cruzi (Luquetti et al., 1999). One year later, Brisse and colleagues (2000) proposed the grouping of T. cruzi isolates into 6 genotypes, or 6 Discrete Typing Units (DTU), named TcI to TcVI (Tc stands for T. cruzi). This is the basis of the current consensus of nomenclature: T. cruzi is a single species composed of 6 distinct DTUs (Zingales et al., 2009).
In addition to these DTUs, in 2009, a new Trypanosoma cruzi genotype, first associated with bats, was described: Tcbat (see Marcili et al., 2009). This genotype is phylogenetically close to TcI but represents a well separated clade. As a T. cruzi parasite, Tcbat is able to infect mice in experimental conditions. Although, its development in triatomines is more efficient in cave triatomines, which is the same pattern observed for another trypanosome associated with bats, T. c. marinkellei (See Marinkelle, 1982). Although first associated with bats, human infections by Tcbat have been reported in a Chilean mummy (Guhl et al., 2014) and a Colombian patient (Ramírez et al., 2014).
The currently most-accepted hypothesis to explain the establishment of the 6 DTUs points to TcI and TcII as the 2 ancestor lineages, and the emergence of the DTUs TcIII and TcIV as a result of a first hybridization process between them. A second and more recent hybridization process involving TcII and TcIII would have given rise to the current hybrid lineages TcV and TcVI (Westenberger et al., 2005).
The geographic distribution of Trypanosoma cruzi DTUs is not completely known and further knowledge of both the geographical and host-expanding data for each DTU will certainly result in distinct distribution maps (Jansen et al., 2015; 2018). However, some aspects can be highlighted: 1) TcI is the most dispersed DTU throughout the Americas; 2) outside South America, mainly TcI and TcIV are detected; 3) TcII occurs in the wild, but in restricted transmission foci and is the genotype associated with human infections in the former endemic areas of the disease (Jansen et al., 2015; 2018); 4) TcIII and TcIV are commonly found in wild mammals and the latter has been associated with oral outbreaks in the Amazon region (Santana et al., 2019); 5) TcV and TcVI are rarer than the others, both in humans and wild mammals; and 6) the subdividing genotypes of the T. cruzi population, previously known as zymodeme 2 (TCII in the first nomenclature consensus), are more recent. Because of that, ancient descriptions of TCI are still considered TcI, but TCII was subdivided into the other 5 subpopulations. This means that, except for TcI, all other T. cruzi DTUs are actually underestimated on the current distribution maps.
The widespread transmission of Trypanosoma cruzi to humans, which led to hundreds of thousands cases per year in the last century, was directly associated with the presence of the domiciliary bug Triatoma infestans (See Figure 6). This bug species originates from the Bolivian valleys of the eastern Andes, where it is found both in domiciles and in wild environments (Panzera et al., 2014). The domiciliary process of Tri. infestans occurred concurrently with the beginning of human settlements near forest environments. At the same time, the process of domestication of local guinea pigs near residences attracted the insects to the home environment (Aufderheide et al., 2004). After the colonization of the Americas by Europeans, the greater movement of people and materials favored the establishment of the intradomiciliary colonies of Tri. infestans, first in Bolivia and subsequently in other countries from South America, especially in those regions south of the Amazon basin. The presence of infected domiciliary bugs in stick houses was what Carlos Chagas found when he described the parasite and the associated vector (Chagas, 1909).
This classical scenario of transmission started to change in the 1970s, with insecticide campaigns to eliminate domiciliary colonies of this kissing bug species (Dias, 2007). In 1991, the governments of Argentina, Bolivia, Brazil, Chile, Paraguay, and Uruguay created the Southern Cone Initiative aiming to eliminate the intradomiciliary transmission of Try. cruzi by Tri. infestans, which was achieved in 2006 (Dias, 2007). The certified interruption of this method of transmission, however, did not represent the end of contaminative vectorial transmission. This manner of transmission continues to occur, but now in the extradomiciliar environment, when humans expose themselves to wild transmission cycles, or in the intradomiciliar environment, usually associated with the invasion of wild vectors and not to domiciliary colonies (Dias et al., 2016). In both cases, control measures adopted against Tri. infestans are not effective. With the virtual elimination of Tri. infestans in some countries, including Brazil, the majority of the new reported cases are concentrated in the Amazon region, a previously considered non-endemic area due to the absence of Tri. infestans in the area. Only in Brazil, approximately 150–200 new cases of Chagas disease are reported per year (Dias et al., 2016). These cases are always associated with infected sylvatic kissing bugs that came in contact with humans.
These sylvatic bugs are immersed in a huge net of Trypanosoma cruzi transmission along with mammals from distinct orders. This transmission net resembles the trophic energy characteristic of food webs (Herrera et al., 2011). Both the oral route, through the predation of mammals and insects, and contaminative transmission are involved in the dispersion of this parasite in nature. Triatomines can be preyed upon by smaller mammals, and these by medium or top-chain carnivores (Herrera et al., 2011; Rocha et al., 2013). Moreover, the different mammal hosts are distributed in distinct forest strata and enable the parasite transmission in the most diverse habitats. This is especially important for mammals with generalist habits, like coatis, opossums, and felids, which frequent different forest strata and, thus, can connect parasite transmission cycles from different environments (Jansen et al., 2018).
Trypanosoma cruzi transmission can occur at all levels of the food web, and each group of mammals also presents varying importance as a reservoir of this parasite. In the pyramid base of consumers are the small terrestrial mammals. These mammals are excellent models of study because they comprise large populations and are usually the group of mammals with the highest biomass in any ecotope (Mills and Childs, 1998). In addition, they have a short lifespan and fast generational turnover, which allows the early identification of environmental changes. In the second level of the pyramid are the bats and medium-sized carnivores. They are classified as mesopredators, since they may be exposed to T. cruzi infection by predation of both infected vectors and small mammals. They are usually generalists and capable of exploiting distinct forest strata. At the top of the pyramid are those animals at the top of the food chain. They are mammals characterized by a wide geographic range and capacity for displacement, which are important conditions for parasite dispersion. In addition, they are considered bioaccumulators of orally transmitted parasites, as is the case for T. cruzi (see Herrera et al., 2011; Rocha et al., 2013).
Small mammals are excellent models of study for identifying spatial and temporal variation in Trypanosoma cruzi transmission. Concerning the spatial differences, the effect of habitat fragmentation on T. cruzi transmission among small mammals has been demonstrated by Vaz et al. (2007). Fragments of forest patches of different sizes (small, medium, and large) were investigated, in addition to a national park, as a preserved control area. Vaz and others (2007) observed that the greater the fragmentation of the wild environment, the lower the diversity of small placental mammals and the greater the abundance of marsupials. This different faunal composition was reflected in distinct infection prevalences, especially in the medium and large fragments (Vaz et al., 2007). Temporal differences were observed in the evaluation of infection prevalences in small mammals from the same locality (Jaguaruana, Ceará State, Brazil) during a 4-year follow-up. The anthropogenic devastation of the area resulted in lower species richness of placentals and a higher abundance of opossums. The consequence for T. cruzi transmission was an increase from an initial rate of 10% of the mammals infected to a rate higher than 50% after the fourth year (Jansen, unpublished data).
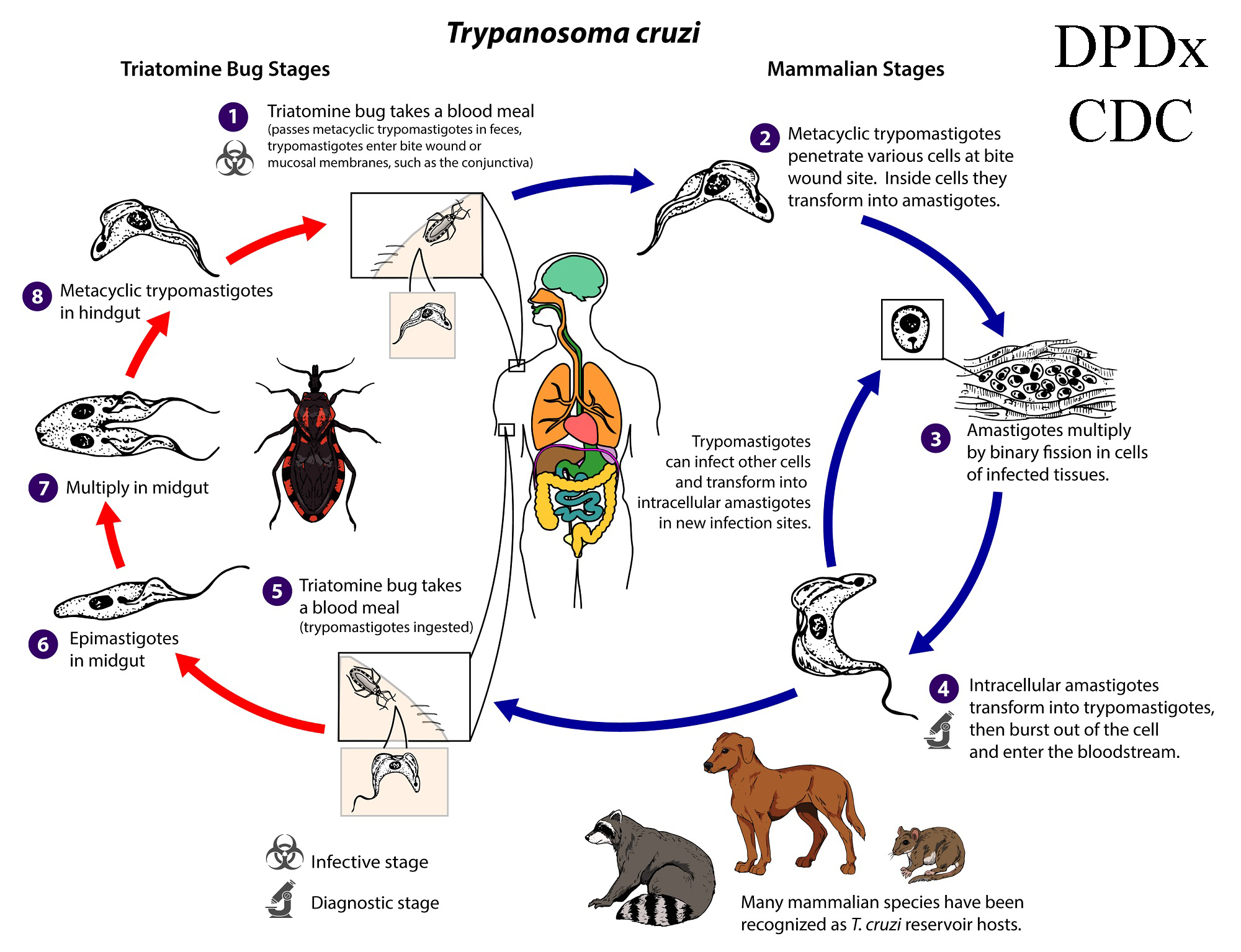
Figure 6. American Trypanosomiasis. Trypanosoma cruzi, is a parasitic protozoan that is the causative agent of Chagas disease (American trypanosomiasis). Currently, 6 distinct lineages of T. cruzi are classified into discrete typing units (TcI–VI), which vary in their geographic occurrence, host specificity, and pathogenicity. Life cycle diagram: An infected triatomine insect vector (or kissing bug) takes a blood meal and releases trypomastigotes in its feces near the site of the bite wound. Trypomastigotes enter the host through the bite wound or intact mucosal membranes, such as the conjunctiva (1). Inside the host, the trypomastigotes invade cells near the site of inoculation, where they differentiate into intracellular amastigotes (2). The amastigotes multiply by binary fission (3) and differentiate into trypomastigotes and then are released into the circulation as bloodstream trypomastigotes (4). Trypomastigotes infect cells from a variety of tissues and transform into intracellular amastigotes in new infection sites. Clinical manifestations can result from this infective cycle. The bloodstream trypomastigotes do not replicate (different from the African trypanosomes). Replication resumes only when the parasites enter another cell or are ingested by another vector. The kissing bug becomes infected by feeding on human or animal blood that contains circulating parasites (5). The ingested trypomastigotes transform into epimastigotes in the vector’s midgut (6). The parasites multiply and differentiate in the midgut (7) and differentiate into infective metacyclic trypomastigotes in the hindgut (8). Other less common routes of transmission include blood transfusions, organ transplantation, transplacental transmission, and foodborne transmission (via food or drink contaminated with the vector and/or its feces).
(Source: United States Centers for Disease Control and Prevention, Global Health, Division of Parasitic Diseases and Malaria, 2021. Public domain.)
An example of a mesopredator that, throughout several years of study, demonstrated to be an important Trypanosoma cruzi reservoir is the coati (Nasua nasua) from the Pantanal region of Brazil. Distinct T. cruzi DTUs (TcI–TcIV) have been found in single mixed infections and long-term follow-up showed that coatis may present high and long-lasting parasitemias that are influenced by seasonality and gender (Herrera et al., 2008; 2011; Alves et al., 2016; Jansen et al., 2018). Coatis nest in very tall trees and their nests serve as microhabitats for several other mammals and insects, including kissing bugs that have been found to be infected by T. cruzi (see De Lima et al., 2015).
Golden lion tamarins—GLT (Leonthopitecus rosalia)—are also demonstrated to be competent Trypanosoma cruzi reservoirs (Lisboa et al., 2015). This callitrichid is an endangered species restricted to the Atlantic rainforest of Rio de Janeiro State, Brazil. It was during the GLT survey for T. cruzi that the presence of TcII (the DTU until then related only to human infections), was observed for the first time in the wild (Lisboa et al., 2000). An 11-year follow-up demonstrated that individuals of L. rosalia are able to maintain long-lasting infections and infectivity potential when infected by T. cruzi from both DTUs TcI and TcII (Lisboa et al., 2015).
Top-predator mammals are still poorly studied, mainly due to the difficulty in trapping and handling them, but are known as important bioaccumulators of Trypanosoma cruzi. In this sense, a positive correlation between the proportion of insects or other mammals in the diet and the infection rates by T. cruzi has been demonstrated (Rocha et al., 2013).
The composition of the local fauna may indicate which Trypanosoma cruzi DTUs will predominate in an area. Didelphis species, for example, are known to maintain high and long-lasting parasitemias (as expressed by positive blood cultures), but only when infected by DTU TcI (Legey et al., 2003), while GLTs can maintain stable infections also by TcII (Lisboa et al., 2015). In contrast, wild and domestic canids usually exhibit a short period of high parasitemia, exactly as observed for humans (Jansen et al., 2018). This diversity explains the differences in the enzootic cycles of the parasite in different areas. In fact, each area is unique and has its own particularities (Jansen et al., 2015).
Currently, what will impact in the infection rates that have emerged in recent outbreaks of Chagas disease in the Americas include: 1) Control of domiciliary transmission of Trypanosoma cruzi by Triatoma infestans; 2) control of the parasite in the wild fauna of vectors and mammals; and 3) environmental disturbances that result in faunal modifications. With these in mind, and aiming to identify common risk factors that could be used in surveillance programs, domestic dogs were proposed as sentinels of imminent risk of transmission to humans (Roque and Jansen, 2008). In these outbreak areas, where a well-established wild transmission cycle of Try. cruzi was observed, dogs showed high prevalence of infection as diagnosed by serology, but did not contribute to the amplification of the parasite population, as attested by negative hemocultures (Roque et al., 2008; 2013). This proposal was further confirmed by spatial analysis using interpolation and map algebra tools, which also considered the environmental factors that interfered with the transmission. In this study, the authors confirmed the hypothesis that loss of wildlife richness was associated with higher parasitemias in wild fauna and higher serological prevalence in the associated canine population (Xavier et al., 2012).
In fact, the study of a multi-host zoonotic parasite such as Trypanosoma cruzi encourages analysis of the transmission to humans using a One Health approach (Thompson, 2013). Additionaly, implementation of the DAMA protocol is urgently needed in order to understand and take action in these and other parasites (Brooks et al., 2014; 2019). Parasite spillover or more properly known as ecological fitting (Janzen, 1985) is a phenomenon frequently amplified by changes in the landscape that: 1) Result in selection of individuals; 2) change the dynamics of transmission; and 3) in the case of zoonosis, will ultimately result in higher or lesser risk of emergence of human cases. Multidisciplinary studies are essential to better understand T. cruzi control and transmission, which are key conditions for successful surveillance programs. As a primarily enzootic parasite, T. cruzi transmission will never be eliminated, but prediction of new human cases and outbreaks is possible and urgently needed.
Literature Cited
Alves, F. M., J. S. de Lima, F. L. Rocha, H. M. Herrera, et al. 2016. Complexity and multi-factoriality of Trypanosoma cruzi sylvatic cycle in coatis, Nasua nasua (Procyonidae), and triatomine bugs in the Brazilian Pantanal. Parasites and Vectors 9: 378. doi: 10.1186/s13071-016-1649-4
Andrade, S. G., M. L. Carvalho, and R. M. Figueira. 1970. Caracterização morfo-biológica e histopatológica de diferentes cepas do Trypanosoma cruzi. Gazeta Médica da Bahia 70: 245–250. https://www.arca.fiocruz.br/handle/icict/17867
Añez, N. 1982. Studies on Trypanosoma rangeli Tejera, 1920, IV: A reconsideration of its systematic position. Memórias do Instituto Oswaldo Cruz 77: 405–415. doi: 10.1590/S0074-02761982000400007
Araújo, A., A. M. Jansen, F. Bouchet, and K. Reinhard. 2003. Parasitism, the diversity of life, and paleoparasitology. Memórias do Instituto Oswaldo Cruz 98 (Supplement 1): 5–11. https://www.arca.fiocruz.br/handle/icict/35746
Aregawi, W. G., G. E. Agga, R. D. Abdi, and P. Büscher. 2019. Systematic review and meta-analysis on the global distribution, host range, and prevalence of Trypanosoma evansi. Parasites and Vectors 12: 67. doi: 10.1186/s13071-019-3311-4
Aufderheide, A. C., W. Salo, M. Madden, J. Streitz, et al. 2004. A 9,000-year record of Chagas’ disease. Proceedings of the National Academy of Sciences of the United States of America 101: 2,034–2,039. doi: 10.1073/pnas.0307312101
Barisón, M. J., L. N. Rapado, E. F. Merino, E. M. Furusho Pral, et al. 2017. Metabolomic profiling reveals a finely tuned, starvation-induced metabolic switch in Trypanosoma cruzi epimastigotes. Journal of Biological Chemistry 292: 8,964–8,977. doi: 10.1074/jbc.M117.778522
Barry, J. D., S. L. Hajduk, K. Vickerman, and D. Le Ray. 1979. Detection of multiple variable antigen types in metacyclic populations of Trypanosoma brucei. Transactions of the Royal Society of Tropical Medicine and Hygiene 73: 205–208. doi: 10.1016/0035-9203(79)90213-X
Bastos, T. S. A., A. M. Faria, D. M. C. Madrid, L. C. Bessa, et al. 2017. First outbreak and subsequent cases of Trypanosoma vivax in the state of Goiás, Brazil. Revista Brasileira de Parasitologia Veterinária 26: 366–371. doi: 10.1590/S1984-29612017019
Batista, J. S., A. F. Oliveira, C. M. Rodrigues, C. A. Damasceno, et al. 2009. Infection by Trypanosoma vivax in goats and sheep in the Brazilian semiarid region: From acute disease outbreak to chronic cryptic infection. Veterinary Parasitology 165: 131–135. doi: 10.1016/j.vetpar.2009.07.005
Batista, J. S., F. Riet-Correa, M. M. Teixeira, C. R. Madruga, et al. 2007. Trypanosomiasis by Trypanosoma vivax in cattle in the Brazilian semiarid: Description of an outbreak and lesions in the nervous system. Veterinary Parasitology 143: 174–181. doi: 10.1016/j.vetpar.2006.08.017
Borghesan, T. C., R. C. Ferreira, C. S. A. Takata, M. Campaner, et al. 2013. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist 164: 129–152. doi: 10.1016/j.protis.2012.06.001
Böse, R., and K. Petersen. 1991. Lipoptena cervi (Diptera), a potential vector of Megatrypanum trypanosomes of deer (Cervidae). Parasitology Research 77: 723–725. doi: 10.1007/BF00928691
Botero, A., C. Cooper, C. K. Thompson, P. L. Clode, et al. 2016. Morphological and phylogenetic description of Trypanosoma noyesi sp. nov.: An Australian wildlife trypanosome within the T. cruzi clade. Protist 167: 425–439. doi: 10.1016/j.protis.2016.07.002
Bourgeon, L., A. Burke, and T. Higham. 2017. Earliest human presence in North America dated to the Last Glacial Maximum: New radiocarbon dates from Bluefish Caves, Canada. PLoS One 12: e0169486. doi: 10.1371/journal.pone.0169486
Brisse, S., J. C. Dujardin, and M. Tibayrenc. 2000. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Molecular and Biochemical Parasitology 111: 95–105. doi: 10.1016/S0166-6851(00)00302-9
Brooks, D. R., E. P. Hoberg, W. A. Boeger, S. L. Gardner, et al. 2014. Finding them before they find us: Informatics, parasites, and environments in accelerating climate change. Comparative Parasitology 81: 155–164. doi: 10.1654/4724b.1
Brooks, D. R., E. P. Hoberg, and W. A. Boeger. 2019. The Stockholm Paradigm. University of Chicago Press, Chicago, Illinois, United States, 400 p. doi: 10.7208/9780226632582
Brun, R., H. Hecker, and Z. R. Lun. 1998. Trypanosoma evansi and T. equiperdum: Distribution, biology, treatment and phylogenetic relationship (a review). Veterinary Parasitology. 79: 95-107. doi: 10.1016/s0304-4017(98)00146-0
Büscher, P., G. Cecchi, V. Jamonneau, and G. Priotto. 2017. Human African trypanosomiasis. Lancet 390: 2,397–2,409. doi: 10.1016/S0140-6736(17)31510-6
Capewell, P., A. Cooper, C. Clucas, W. Weir, et al. 2015. A co-evolutionary arms race: Trypanosomes shaping the human genome, humans shaping the trypanosome genome. Parasitology 142 (Supplement 1): S108–S119. doi: 10.1017/S0031182014000602
Carnes, J., A. Anupama, O. Balmer, A. Jackson, et al. 2015. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Neglected Tropical Diseases 9: e3404. doi: 10.1371/journal.pntd.0003404
Cecchi, G., M. Paone, R. Argilés Herrero, M. J. Vreysen, et al. 2015. Developing a continental atlas of the distribution and trypanosomal infection of tsetse flies (Glossina species). Parasites and Vectors 8: 284. doi: 10.1186/s13071-015-0898-y
Chagas, C. 1909. Nova tripanozomiase humana: Estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Memórias do Instituto Oswaldo Cruz 1: 159–218. https://www.biodiversitylibrary.org/part/150058
Coura, J. R. 2015. The main sceneries of Chagas’ disease transmission, the vectors, blood and oral transmissions: A comprehensive review. Memórias do Instituto Oswaldo Cruz 110: 277–282. doi: 10.1590/0074-0276140362
Dario, M. A., C. V. Lisboa, L. M. Costa, R. Moratelli, et al. 2017a. High Trypanosoma spp. diversity is maintained by bats and triatomines in Espírito Santo state, Brazil. PLoS One 12: e0188412. doi: 10.1371/journal.pone.0188412
Dario, M. A., R. Moratelli, P. Schwabl, A. M. Jansen, et al. 2017b. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Neglected Tropical Diseases 11: e0005790. doi: 10.1371/journal.pntd.0005790
Dario, M. A., M. S. Rodrigues, J. H. Barros, S. C. Xavier, et al. 2016. Ecological scenario and Trypanosoma cruzi DTU characterization of a fatal acute Chagas disease case transmitted orally (Espírito Santo State, Brazil). Parasites and Vectors 9: 477. doi: 10.1186/s13071-016-1754-4
Da Silva, A. S., H. A. García Pérez, M. M. Costa, R. T. França, et al. 2011. Horses naturally infected by Trypanosoma vivax in southern Brazil. Parasitology Research 108: 23–30. doi: 10.1007/s00436-010-2036-2
D’Avila-Levy, C. M., C. Boucinha, A. Kostygov, H. L. C. Santos, et al. 2015. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Memórias do Instituto Oswaldo Cruz 110: 956–965. doi:10.1590/0074-02760150253
Deane, L. M., M. P. Deane, and R. Lourenço-de-Oliveira. 1986. Are Asian monkeys the original mammalian hosts of Trypanosoma conorhini? Memórias do Instituto Oswaldo Cruz 81: 127–129. doi: 10.1590/S0074-02761986000100018
Deane, M. P., H. L. Lenzi, and A. M. Jansen. 1984. Trypanosoma cruzi: Vertebrate and invertebrate cycles in the same mammal host, the opossum Didelphis marsupialis. Memórias do Instituto Oswaldo Cruz 79: 513–515. doi: 10.1590/S0074-02761984000400021
De Araújo, V. A., M. C. Boité, E. Cupolillo, A. M. Jansen, et al. 2013. Mixed infection in the anteater Tamandua tetradactyla (Mammalia: Pilosa) from Pará State, Brazil: Trypanosoma cruzi, T. rangeli, and Leishmania infantum. Parasitology 140:455–460. doi: 10.1017/S0031182012001886
De Lima, J. S., F. L. Rocha, F. M. Alves, E. S. Lorosa, et al.2015. Infestation of arboreal nests of coatis by triatomine species, vectors of Trypanosoma cruzi, in a large Neotropical wetland. Journal of Vector Ecology 40: 379–385. doi: 10.1111/jvec.12177
De Sousa, M. A. 2014. On opportunist infections by Trypanosoma lewisi in humans and its differential diagnosis from T. cruzi and T. rangeli. Parasitology Research 113: 4,471–4,475. doi: 10.1007/s00436-014-4132-1
De Sousa, M. A., T. da Silva Fonseca, B. N. Dos Santos, S. M.Dos Santos Pereira, et al. 2008. Trypanosoma rangeli Tejera, 1920, in chronic Chagas’ disease patients under ambulatory care at the Evandro Chagas Clinical Research Institute (IPEC-Fiocruz, Brazil). Parasitology Research 103: 697–703. doi: 10.1007/s00436-008-1033-1
Desquesnes, M., A. Dargantes, L. De-Hua, Z. R. Lun, et al. 2013a. Trypanosoma evansi and Surra: A review and perspectives on transmission, epidemiology and control, impact, and zoonotic aspects. Biomed Research International 2013: 321237. doi: 10.1155/2013/321237
Desquesnes, M., P. Holzmuller, L. De-Hua, A. Dargantes, et al. 2013b. Trypanosoma evansi and Surra: A review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Research International 2013: 194176. doi: 10.1155/2013/194176
Dias, J. C. P. 2007. Southern Cone Initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusional Chagas disease: Historical aspects, present situation, and perspectives. Memórias do Instituto Oswaldo Cruz 102 (Supplement 1): 11–18. doi: 10.1590/S0074-02762007005000092
Dias, J. C., A. N. Ramos, Jr., E. D. Gontijo, A. Luquetti, et al. 2016. 2nd Brazilian Consensus on Chagas Disease, 2015. Revista da Sociedade Brasileira de Medicina Tropical 49 (Supplement 1): 3–60. doi: 10.1590/0037-8682-0505-2016
Dos Santos, F. C. B., C. V. Lisboa, S. C. C. Xavier, M. A. Dario, et al. 2018. Trypanosoma sp. diversity in Amazonian bats (Chiroptera; Mammalia) from Acre State, Brazil. Parasitology 145: 828–837. doi: 10.1017/S0031182017001834
Drew, P. A., and C. R. Jenkin. 1982. Properties of ablastin, a factor in the serum of rats infected with Trypanosoma lewisi which inhibits the parasites’ division. Australian Journal of Experimental Biology and Medical Science 60: 329–337. doi: 10.1038/icb.1982.36
Dyer, N. A., C. Rose, N. O. Ejeh, and A. Acosta-Serrano. 2013. Flying tryps: Survival and maturation of trypanosomes in tsetse flies. Trends in Parasitology 29: 188–196. doi: 10.1016/j.pt.2013.02.003
Espinosa, O. A., M. G. Serrano, E. P. Camargo, M. M. G. Teixeira, et al. 2018. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145: 430–442. doi: 10.1017/S0031182016002092
Ferreira, L. L., M. G. Lorenzo, S. L. Elliot, and A. A. Guarneri. 2010. A standardizable protocol for infection of Rhodnius prolixus with Trypanosoma rangeli, which mimics natural infections and reveals physiological effects of infection upon the insect. Journal of Invertebrate Pathology 105: 91–97. doi: 10.1016/j.jip.2010.05.013
Fetene, E., S. Leta, F. Regassa, P. Büscher. 2021. Global distribution, host range, and prevalence of Trypanosoma vivax: A systematic review and meta-analysis. Parasites and Vectors 14: 80. doi: 10.1186/s13071-021-04584-x
Flynn, J. J., and A. R. Wyss. 1998. Recent advances in South American mammalian paleontology. Tree 13: 449–454. doi: 10.1016/S0169-5347(98)01457-8
Ganyo, E. Y., J. N. Boampong, D. K. Masiga, J. Villinger, et al. 2018. Haematology of N’Dama and West African shorthorn cattle herds under natural Trypanosoma vivax challenge in Ghana. F1000 Research 7: 314. doi: 10.12688/f1000research.14032.2
García, E. S., D. P. Castro, M. B. Figueiredo, and P. Azambuja. 2012. Parasite-mediated interactions within the insect vector: Trypanosoma rangeli strategies. Parasites and Vectors 5: 105. doi: 10.1186/1756-3305-5-105
García, H. A., K. Kamyingkird, A. C. Rodrigues, and S. Jittapalapong, et al. 2011. High genetic diversity in field isolates of Trypanosoma theileri assessed by analysis of cathepsin L-like sequences disclosed multiple and new genotypes infecting cattle in Thailand. Veterinary Parasitology 180: 363–367. doi: 10.1016/j.vetpar.2011.03.017
Gardner, S. L., and M. L. Campbell. 1992. Parasites as probes for biodiversity. Journal of Parasitology 78: 596–600. doi: 10.2307/3283534
Gascon, J., C. Bern, and M. J. Pinazo. 2010. Chagas disease in Spain, the United States, and other non-endemic countries. Acta Tropica 115: 22–27. doi: 10.1016/j.actatropica.2009.07.019
Ghosh, S., P. Banerjee, A. Sarkar, S. Datta, et al. 2012. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. Journal of Clinical Microbiology 50: 2,774–2,778. doi:10.1128/JCM.00966-12
Gibson, W. 2015. Liaisons dangereuses: Sexual recombination among pathogenic trypanosomes. Research in Microbiology 166: 459–466. doi: 10.1016/j.resmic.2015.05.005
Gibson, W., J. G. Pilkington, and J. M. Pemberton. 2010. Trypanosoma melophagium from the sheep ked Melophagus ovinus on the island of St. Kilda. Parasitology 137: 1,799–1,804. doi: 10.1017/S0031182010000752
Gizaw, Y., M. Megersa, and T. Fayera. 2017. Dourine: A neglected disease of equids. Tropical Animal Health and Production 49: 887–897. doi: 10.1007/s11250-017-1280-1
Grisard, E. C. 2002. Salivaria or Stercoraria? The Trypanosoma rangeli dilemma. Kinetoplastid Biology Disease 1: 5. doi: 10.1186/1475-9292-1-5
Gross, N. T., O. M. Guerrero, M. Chinchilla, and C. Jarstrand-Hall. 2006. Trypanosoma lewisi-induced immunosuppression: The effects on alveolar macrophage activities against Cryptococcus neoformans. Experimental Parasitology 113: 262–266. doi: 10.1016/j.exppara.2006.02.002
Guhl, F., and G. A. Vallejo. 2003. Trypanosoma (Herpetosoma) rangeli Tejera, 1920: An updated review. Memórias do Instituto Oswaldo Cruz 98: 435–442. doi: 10.1590/S0074-02762003000400001
Guhl, F., A. Aufderheide, and J. D. Ramírez. 2014. From ancient to contemporary molecular eco-epidemiology of Chagas disease in the Americas. International Journal for Parasitology 44: 605–612. doi: 10.1016/j.ijpara.2014.02.005
Hamill, L. C., M. T. Kaare, S. C. Welburn, and K. Picozzi. 2013. Domestic pigs as potential reservoirs of human and animal trypanosomiasis in northern Tanzania. Parasites and Vectors 6: 322. doi: 10.1186/1756-3305-6-322
Hamilton, P. B., E. R. Adams, F. Njiokou, W. C. Gibson, et al. 2009. Phylogenetic analysis reveals the presence of the Trypanosoma cruzi clade in African terrestrial mammals. Infection, Genetics, and Evolution 9: 81–86. doi: 10.1016/j.meegid.2008.10.011
Hamilton, P. B., J. R. Stevens, P. Holz, B. Boag, et al. 2005. The inadvertent introduction into Australia of Trypanosoma nabiasi, the trypanosome of the European rabbit (Oryctolagus cuniculus), and its potential for biocontrol. Molecular Ecology 14: 3,167–3,175. doi: 10.1111/j.1365-294X.2005.02602.x
Hamilton, P. B., M. M. Teixeira, and J. R. Stevens. 2012. The evolution of Trypanosoma cruzi: The ‘bat seeding’ hypothesis. Trends in Parasitology 28: 136–141. doi: 10.1016/j.pt.2012.01.006
Herrera, H. M., A. C. Alessi, L. C. Marques, A. E. Santana, et al. 2002. Experimental Trypanosoma evansi infection in South American coati (Nasua nasua): Hematological, biochemical and histopathological changes. Acta Tropica 81: 203–210. doi: 10.1016/S0001-706X(01)00204-2
Herrera, H. M., A. M. R. D’Ávila, A. Norek, U. G. Abreu, et al. 2004. Enzootiology of Trypanosoma evansi in Pantanal, Brazil. Veterinary Parasitology 125: 263–275. doi: 10.1016/j.vetpar.2004.07.013
Herrera, H. M., C. V. Lisboa, A. P. Pinho, and N. Olifiers. 2008. The coati (Nasua nasua, Carnivora: Procyonidae) as a reservoir host for the main lineages of Trypanosoma cruzi in the Pantanal region, Brazil. Transactions of the Royal Society of Tropical Medicina and Hygiene 102: 1,133–1,139. doi: 10.1016/j.trstmh.2008.04.041
Herrera, H. M., F. L. Rocha, C. V. Lisboa, V. Rademaker, et al. 2011. Food web connections and the transmission cycles of Trypanosoma cruzi and Trypanosoma evansi (Kinetoplastida: Trypanosomatidae) in the Pantanal Region, Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene 105: 380–387. doi: 10.1016/j.trstmh.2011.04.008
Hoare, C. A. 1972. The Trypanosomes of Mammals: A Zoological Monograph. Blackwell Scientific, Oxford, United Kingdom, 750 p.
Hodo, C. L., C. C. Goodwin, B. C. Mayes, J. A. Mariscal, et al. 2016. Trypanosome species, including Trypanosoma cruzi, in sylvatic and peridomestic bats of Texas, USA. Acta Tropica 164: 259–266. doi: 10.1016/j.actatropica.2016.09.013
Jaimes-Dueñez, J., O. Triana-Chávez, A. Valencia-Hernández, D. Sánchez-Arévalo, et al. 2017. Molecular diagnosis and phylogeographic analysis of Trypanosoma evansi in dogs (Canis lupus familiaris) suggest an epidemiological importance of this species in Colombia. Preventive Veterinary Medicine 139: 82–89. doi: 10.1016/j.prevetmed.2017.02.007
Jansen, A. M., S. C. C. Xavier, and A. L. R. Roque. 2015. The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Tropica 151: 1–15. doi: 10.1016/j.actatropica.2015.07.018
Jansen, A. M., S. C. C. Xavier, and A. L. R. Roque. 2018. Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil. Parasites and Vectors 11: 502. doi: 10.1186/s13071-018-3067-2
Janzen, D. H. 1985. On ecological fitting. Oikos 45: 308–310. doi: 10.2307/3565565
Jenkins, E. J., A. Simon, N. Bachand, and C. Stephen. 2015. Wildlife parasites in a One Health world. Trends in Parasitology 31: 174–180. doi: 10.1016/j.pt.2015.01.002
Kaufer, A., J. Ellis, D. Stark, and J. Barratt. 2017. The evolution of trypanosomatid taxonomy. Parasites and Vectors 10: 287. doi: 10.1186/s13071-017-2204-7
Kelly, S., A. Ivens, G. A. Mott, E. O’Neill, et al. 2017. An alternative strategy for trypanosome survival in the mammalian bloodstream revealed through genome and transcriptome analysis of the ubiquitous bovine parasite Trypanosoma (Megatrypanum) theileri. Genome Biology and Evolution 9: 2,093–2,109. doi: 10.1093/gbe/evx152
Kessler, R. L., V. T. Contreras, N. P. Marliére, A. Aparecida Guarneri, et al. 2017. Recently differentiated epimastigotes from Trypanosoma cruzi are infective to the mammalian host. Molecular Microbiology 104: 712–736. doi: 10.1111/mmi.13653
Kingston, N. 1991. A brief review of Trypanosoma (Megatrypanum) infections in ruminants in North America and Europe. Wiadomości parazytologiczne 37: 211–218.
Kolev, N. G., A. Günzl, and C. Tschudi. 2017. Metacyclic VSG expression site promoters are recognized by the same general transcription factor that is required for RNA polymerase I transcription of bloodstream expression sites. Molecular Biochemical Parasitology 216: 52–55. doi: 10.1016/j.molbiopara.2017.07.002
Kovářová, J., R. Nagar, J. Faria, M. A. J. Ferguson, et al. 2018. Gluconeogenesis using glycerol as a substrate in bloodstream-form Trypanosoma brucei. PLoS Pathogens 14: e1007475. doi: 10.1371/journal.ppat.1007475
Latif, A. A., M. A. Bakheit, A. E. Mohamed, and E. Zweygarth. 2004. High infection rates of the tick Hyalomma anatolicum anatolicum with Trypanosoma theileri. Onderstepoort Journal of Veterinary Research 71: 251–256.
Lavocat, R. 1974. Interrelationships between African and South American rodents and their bearing on problem of origin of South American monkeys. Journal of Human Evolution 3: 323–326. doi: 10.1016/0047-2484(74)90027-X
Lee, Y.-F., C.-C. Cheng, J.-S. Chen, N.-N. Lin, et al. 2013. Evidence of intracellular stages in Trypanosoma (Megatrypanum) theileri in non-phagocytic mammalian cells. Veterinary Parasitology 191: 228–239. doi: 10.1016/j.vetpar.2012.08.027
Legey, A. P., A. P. Pinho, S. C. C. Xavier, R. Marchevsky, et al. 2003. Trypanosoma cruzi in marsupial didelphids (Philander frenata and Didelphis marsupialis): Differences in the humoral immune response in natural and experimental infections. Revista da Sociedade Brasileira de Medicina Tropical 36: 241–248. doi: 10.1590/S0037-86822003000200008
Leonard, G., D. M. Soanes, and J. R. Stevens. 2011. Resolving the question of trypanosome monophyly: A comparative genomics approach using whole genome data sets with low taxon sampling. Infection, Genetics, and Evolution 11: 955–959. doi: 10.1016/j.meegid.2011.03.005
Lewis, M. D., M. S. Llewellyn, M. Yeo, N. Acosta, et al. 2011. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Neglected Tropical Diseases 5: e1363. doi: 10.1371/journal.pntd.0001363
Lima, L., O. Espinosa-Álvarez, P. B. Hamilton, L. Neves, et al. 2013. Trypanosoma livingstonei: A new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasites and Vectors 6: 221. doi: 10.1186/1756-3305-6-221
Lima, L., O. Espinosa-Álvarez, P. A. Ortiz, J. A. Trejo-Varón, et al. 2015a. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Tropica 151: 166–177. doi: 10.1016/j.actatropica.2015.07.015
Lima, L., O. Espinosa-Álvarez, C. M. Pinto, and M. Cavazzana, Jr. 2015b. New insights into the evolution of the Trypanosoma cruzi clade provided by a new trypanosome species tightly linked to Neotropical Pteronotus bats and related to an Australian lineage of trypanosomes. Parasites and Vectors 8: 657. doi: 10.1186/s13071-015-1255-x
Lima, L., F. M. Silva, L. Neves, M. Attias, et al. 2012. Evolutionary insights from bat trypanosomes: Morphological, developmental, and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist 163: 856–872. doi: 10.1016/j.protis.2011.12.003
Lin, R.-H., D.-H. Lai, L.-L. Zheng, J. Wu, et al. 2015. Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen. Parasites and Vectors 8: 665. doi: 10.1186/s13071-015-1281-8
Lisboa, C. V., J. Dietz, A. J. Baker, N. N. Russel, et al. 2000. Trypanosoma cruzi infection in Leontopithecus rosalia at the Reserva Biológica de Poco das Antas, Rio de Janeiro, Brazil. Memórias do Instituto Oswaldo Cruz 95: 445–452. doi: 10.1590/S0074-02762000000400002
Lisboa, C. V., R. V. Monteiro, A. F. Martins, S. C. C. Xavier, et al. 2015. Infection with Trypanosoma cruzi TcII and TcI in free-ranging population of lion tamarins (Leontopithecus spp): An 11-year follow-up. Memórias do Instituto Oswaldo Cruz 110: 394–402. doi: 10.1590/0074-02760140400
Lizundia, R., C. Newman, C. D. Buesching, D. Ngugi, et al. 2011. Evidence for a role of the host-specific flea (Paraceras melis) in the transmission of Trypanosoma (Megatrypanum) pestanai to the European badger. PLoS One 6: e16977. doi: 10.1371/journal.pone.0016977
Lopes, C. M. T., R. F. S. Menna-Barreto, M. G. Pavan, M. C. S. Pereira, et al. 2018. Trypanosoma janseni n. sp. (Trypanosomatida: Trypanosomatidae) isolated from Didelphis aurita (Mammalia: Didelphidae) in the Atlantic rainforest of Rio de Janeiro, Brazil: Integrative taxonomy and phylogeography within the Trypanosoma cruzi clade. Memórias do Instituto Oswaldo Cruz 113: 45–55. doi: 10.1590/0074-02760170297
Luquetti, A., A. Prata, A. Moncayo, A. Romanha, et al. 1999. Recommendations from a satellite meeting. From the International Symposium to Commemorate the 90th Anniversary of the Discovery of Chagas Disease, April 11–16, 1999, Rio de Janeiro, Brazil. Memórias do Instituto Oswaldo Cruz 94 (Supplement 1): 429–432. https://www.scielo.br/j/mioc/a/q9RgPTFzjvjgyWL7cvf3WJs/?lang=en&format=pdf
Mafie, E., A. Saito-Ito, M. Kasai, M. Hatta, et al. 2019. Integrative taxonomic approach of trypanosomes in the blood of rodents and soricids in Asian countries, with the description of three new species. Parasitology Research 118: 97–109. doi: 10.1007/s00436-018-6120-3
Maia da Silva, F., A. Marcili, L. Lima, M. Cavazzana, Jr., et al. 2009. Trypanosoma rangeli isolates of bats from central Brazil: Genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Tropica 109: 199–207. doi: 10.1016/j.actatropica.2008.11.005
Maia da Silva, F., A. Marcili, P. A., Ortiz, S. Epiphanio, et al. 2010. Phylogenetic, morphological, and behavioural analyses support host switching of Trypanosoma (Herpetosoma) lewisi from domestic rats to primates. Infection, Genetics, and Evolution 10: 522–529. doi: 10.1016/j.meegid.2010.02.005
Maraghi, S., K. R. Wallbanks, and D. H. Molyneux. 1995. Oral transmission of trypanosomes of the subgenus Herpetosoma from small mammals. Parasitology Research 81: 693–695.
Marcili, A., L. Lima, M. Cavazzana, A. C. Junqueira, et al. 2009. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology 136: 641–655. doi: 10.1017/S0031182009005861
Marinkelle, C. J. 1982. Developmental stages of Trypanosoma cruzi-like flagellates in Cavernicola pilosa. Revista de Biología Tropical 30: 107–111.
Miles, M. A., P. J. Toyé, S. C. Oswald, and D. G. Godfrey. 1977. The identification by isoenzyme patterns of two distinct strains groups of Trypanosoma cruzi circulating independently in a rural area of Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene 71: 217–225. doi: 10.1016/0035-9203(77)90012-8
Mills, J. N., and J. E. Childs. 1998. Ecologic studies of rodent reservoirs: Their relevance of human health. Emerging Infectious Diseases 4: 529–537. doi: 10.3201/eid0404.980403
Monroy, F. P., and D. G. Dusanic. 2000. The kidney form of Trypanosoma musculi: A distinct stage in the life cycle? Parasitology Today 16: 107–110. doi: 10.1016/S0169-4758(99)01599-9
Moreira, D., P. López-García, and K. Vickerman. 2004. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: Proposal for a new classification of the class Kinetoplastea. International Journal of Systematic and Evolutionary Microbiology 54: 1,861–1,875. doi: 10.1099/ijs.0.63081-0
Morio, F., J. Reynes, M. Dollet, F. Pratlong, et al. 2008. Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. Journal of Clinical Microbiology 46: 3,845–3,847. doi: 10.1128/JCM.01098-08
Morrison, L. J., L. Vezza, T. Rowan, and J. C. Hope. 2016. Animal African trypanosomiasis: Time to increase focus on clinically relevant parasite and host species. Trends in Parasitology 32: 599–607. doi: 10.1016/j.pt.2016.04.012
Mugnier, M. R., C. E. Stebbins, and F. N. Papavasiliou. 2016. Masters of Disguise: Antigenic Variation and the VSG Coat in Trypanosoma brucei. PLoS Pathogens 12: e1005784. doi: 10.1371/journal.ppat.1005784
Osório, A. L., C. R. Madruga, M. Desquesnes, C. O. Soares, et al. 2008. Trypanosoma (Duttonella) vivax, its biology, epidemiology, pathogenesis, and introduction in the New World: A review. Memórias do Instituto Oswaldo Cruz 103: 1–13. doi: 10.1590/S0074-02762008000100001
PAHO (Pan-American Health Organization). 2009. Doença de Chagas: Guia para vigilância, prevenção, controle e manejo clínico da doença de chagas aguda transmitida por alimentos, 92 p. https://bvsms.saude.gov.br/bvs/publicacoes/guia_vigilancia_prevencao_doenca_chagas.pdf
Panzera, F., M. J. Ferreiro, S. Pita, L. Calleros, et al. 2014. Evolutionary and dispersal history of Triatoma infestans, main vector of Chagas disease, by chromosomal markers. Infection, Geneticas and Evolution 27: 105–113. doi: 10.1016/j.meegid.2014.07.006
Peirce, M. A., and C. Neal. 1974. Trypanosoma (Megatrypanum) pestanai in British badgers (Meles meles). International Journal for Parasitology 4: 439–440. doi: 10.1016/0020-7519(74)90055-1
Poulin, R. 2014. Parasite biodiversity revisited: Frontiers and constraints. International Journal for Parasitology 44: 581–589. doi: 10.1016/j.ijpara.2014.02.003
Pronovost, H., A. C. Peterson, B. G. Chavez, M. J. Blum, et al. 2020. Deep sequencing reveals multiclonality and new discrete typing units of Trypanosoma cruzi in rodents from the southern United States. Journal of Microbiology, Immunology, and Infection 53: 622–623. doi: 10.1016/j.jmii.2018.12.004
Rademaker, V., H. M. Herrera, T. R. Raffel, P. S. D’Andrea, et al. 2009. What is the role of small rodents in the transmission cycle of Trypanosoma cruzi and Trypanosoma evansi (Kinetoplastida: Trypanosomatidae)? A study case in the Brazilian Pantanal. Acta Tropica 111: 102–107. doi: 10.1016/j.actatropica.2009.02.006
Radwanska, M., N. Vereecke, V. Deleeuw, J. Pinto, et al. 2018. Salivarian trypanosomosis: A review of parasites involved, their global distribution and their interaction with the innate and adaptive mammalian host immune system. Frontiers in Immunology 9: 2253. doi: 10.3389/fimmu.2018.02253
Ramírez, J. D., C. Hernández, M. Montilla, P. Zambrano, et al. 2014. First report of human Trypanosoma cruzi infection attributed to TcBat genotype. Zoonoses Public Health 61: 477–479. doi: 10.1111/zph.12094
Raven, P. H., and D. I. Axelrod. 1975. History of the flora and fauna of Latin America: The theory of plate tectonics provides a basis for reinterpreting the origins and distribution of the biota. American Scientist 63: 420–429.
Ríos Carrera, N. J., M. C. Chincilla Carmona, O. M. Guerrero, and A. Castro Castillo. 2009. [The immunosuppressant effect of T. lewisi (Kinetoplastidae) infection on the multiplication of Toxoplasma gondii (Sarcocystidae) on alveolar and peritoneal macrophages of the white rat.] Revista de Biología Tropical 57: 13–22. [In Spanish.]
Rocha, F. L., A. L. R. Roque, J. S. de Lima, C. C. Cheida, et al. 2013. Trypanosoma cruzi infection in Neotropical wild carnivores (Mammalia: Carnivora): At the top of the T. cruzi transmission chain. PLoS One 8: e67463. doi: 10.1371/journal.pone.0067463
Rodrigues, A. C., P. A. Ortiz, A. G. Costa-Martins, L. Neves, et al. 2014. Congopain genes diverged to become specific to Savannah, Forest, and Kilifi subgroups of Trypanosoma congolense, and are valuable for diagnosis, genotyping and phylogenetic inferences. Infection, Genetics, and Evolution 23: 20–31. doi: 10.1016/j.meegid.2014.01.012
Rodrigues, C. M., J. S. Batista, J. M. Lima, J. F. Freitas, et al. 2015. Field and experimental symptomless infections support wandering donkeys as healthy carriers of Trypanosoma vivax in the Brazilian Semiarid, a region of outbreaks of high mortality in cattle and sheep. Parasites and Vectors 8: 564. doi: 10.1186/s13071-015-1169-7
Romero-Meza, G., and M. R. Mugnier. 2020. Trypanosoma brucei. Trends in Parasitology 36: 571–572. doi: 10.1016/j.pt.2019.10.007
Roque, A. L. R., and A. M. Jansen. 2008. The importance of sentinel domestic animals to identify risk areas to the emergence of Chagas disease. Revista da Sociedade Brasileira de Medicina Tropical 41: 191–193.
Roque, A. L. R., S. C. C. Xavier, M. G. da Rocha, A. C. Duarte, et al. 2008. Trypanosoma cruzi transmission cycle among wild and domestic mammals in three areas of orally transmitted Chagas disease outbreaks. American Journal of Tropical Medicine and Hygiene 79: 742–749.
Roque, A. L. R., S. C. Xavier, M. Gerhardt, M. F. Silva, et al. 2013. Trypanosoma cruzi among wild and domestic mammals in different areas of the Abaetetuba municipality (Pará State, Brazil), an endemic Chagas disease transmission area. Veterinary Parasitology 193: 71–77. doi: 10.1016/j.vetpar.2012.11.028
Sánchez, E., T. Perrone, G. Recchimuzzi, I. Cardozo, et al. 2015. Molecular characterization and classification of Trypanosoma spp. Venezuelan isolates based on microsatellite markers and kinetoplast maxicircle genes. Parasites and Vectors 8: 536. doi: 10.1186/s13071-015-1129-2
Sánchez, E., T. Perrone, G. Recchimuzzi, I. Cardozo, et al. 2015. Molecular characterization and classification of Trypanosoma spp. Venezuelan isolates based on microsatellite markers and kinetoplast maxicircle genes [Erratum]. Parasites and Vectors 8: 566. doi: 10.1186/s13071-015-1177-7
Santana, R. A. G., M. G. V. B. Guerra, D. R. Sousa, K. Couceiro, et al. 2019. Oral transmission of Trypanosoma cruzi, Brazilian Amazon. Emerging Infectious Diseases 25: 132–135. doi: 10.3201/eid2501.180646
Shah, I., U. S. Ali, P. Andankar, and R. R. Joshi. 2011. Trypanosomiasis in an infant from India. Journal of Vector Borne Diseases 48: 122–123.
Sharma, R., E. Gluenz, L. Peacock, W. Gibson, et al. 2009. The heart of darkness: Growth and form of Trypanosoma brucei in the tsetse fly. Trends in Parasitology 25: 517–524. doi: 10.1016/j.pt.2009.08.001
Shaw, J. J., and R. Lainson. 1972. Trypanosoma vivax in Brazil. Annals of Tropical Medicine and Parasitology 66: 25–32.
Shlomai, J. 2004. The structure and replication of kinetoplast DNA. Current Molecular Medicine 4: 623–647. doi: 10.2174/1566524043360096
Silva, R. A., J. A. da Silva, R. C. Schneider, J. de Freitas., et al. 1996. Outbreak of trypanosomiasis due to Trypanosoma vivax (Ziemann, 1905) in bovines of the Pantanal, Brazil. Memórias do Instituto Oswaldo Cruz 91: 561–562. doi: 10.1590/S0074-02761996000500005
Souza, W. 1999. A short review on the morphology of Trypanosoma cruzi: From 1909 to 1999. Memórias do Instituto Oswaldo Cruz 94 (Supplement I): 17–36. doi: 10.1590/S0074-02761999000700003
Stevens, J. R., H. A. Noyes, G. A., Dover, and W. C. Gibson. 1999a. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology 118: 107–116.
Stevens, J. R., H. A. Noyes, C. J. Schofield, and W. Gibson. 2001. The molecular evolution of Trypanosomatidae. Advances in Parasitology 48: 1–56. doi: 10.1016/s0065-308x(01)48003-1
Stevens, J. R., M. M. Teixeira, L. E. Bingle, and W. C. Gibson. 1999b. The taxonomic position and evolutionary relationships of Trypanosoma rangeli. International Journal for Parasitology 29: 749–757. doi: 10.1016/S0020-7519(99)00016-8
Stijlemans, B., G. Caljon, J. Van Den Abbeele, J. A. Van Ginderachter, et al. 2016. Immune evasion strategies of Trypanosoma brucei within the mammalian host: Progression to pathogenicity. Frontiers in Immunology 7: 233. doi: 10.3389/fimmu.2016.00233
Suganuma, K., S. Narantsatsral, B. Battur, S. Yamasaki, et al. 2016. Isolation, cultivation, and molecular characterization of a new Trypanosoma equiperdum strain in Mongolia. Parasites and Vectors 9: 481. doi: 10.1186/s13071-016-1755-3
Thompson, R. C. A. 2013. Parasite zoonoses and wildlife: One Health, spillover, and human activity. International Journal for Parasitology 43: 1,079–1,088. doi: 10.1016/j.ijpara.2013.06.007
Tomlinson, S., A. M. Jansen, A. Koudinov, J. A. Ghiso, et al. 1995. High-density-lipoprotein-independent killing of Trypanosoma brucei by human serum. Molecular Biochemical Parasitology 70: 131–138. doi: 10.1016/0166-6851(95)00019-W
Truc, P. 1996. A miniature kit for the in vitro isolation of Trypanosoma brucei gambiense: A preliminary field assessment on sleeping sickness patients in Côte d’Ivoire. Transactions of the Royal Society of Tropical Medicine and Hygiene 90: 246–247. doi: 10.1016/s0035-9203(96)90232-1
Urrea, D. A., F. Guhl, C. P. Herrera, A. Falla, et al. 2011. Sequence analysis of the spliced-leader intergenic region (SL-IR) and random amplified polymorphic DNA (RAPD) of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis, R. colombiensis, R. pallescens and R. prolixus suggests a degree of co-evolution between parasites and vectors. Acta Tropica 120: 59–66. doi: 10.1016/j.actatropica.2011.05.016
Vallejo, G. A., F. Guhl, J. C. Carranza, L. E. Lozano, et al. 2002. kDNA markers define two major Trypanosoma rangeli lineages in Latin America. Acta Tropica 81: 77–82. doi: 10.1016/S0001-706X(01)00186-3
Van den Bossche, P., R. De Deken, J. Brandt, S. Geerts, et al. 2004. The transmission of mixed Trypanosoma brucei brucei/T. congolense infections by tsetse (Glossina morsitans morsitans). Veterinary Parasitology 119: 147–153. doi: 10.1016/j.vetpar.2003.11.008
Vaz, V. C., P. S. D’Andrea, and A. M. Jansen. 2007. Effects of habitat fragmentation on wild mammal infection by Trypanosoma cruzi. Parasitology 134: 1,785–1,793. doi: 10.1017/S003118200700323X
Verma, A., S. Manchanda, N. Kumar, A. Sharma, et al. 2011. Trypanosoma lewisi or T. lewisi-like infection in a 37-day-old Indian infant. American Journal of Tropical Medicine and Hygiene 85: 221–224. doi: 10.4269/ajtmh.2011.11-0002
Vickerman, K., L. Tetley, K. A. Hendry, and C. M. Turner. 1988. Biology of African trypanosomes in the tsetse fly. Biology of the Cell 64: 109–119. doi: 10.1016/0248-4900(88)90070-6
Wargnies, M., E. Bertiaux, E. Cahoreau, N. Ziebart, et al. 2018. Gluconeogenesis is essential for trypanosome development in the tsetse fly vector. PLoS Pathogens 14: e1007502. doi: 10.1371/journal.ppat.1007502
Wen, Y.-Z., Z.-R. Lun, X.-Q. Zhu, G. Hide, et al. 2016. Further evidence from SSCP and ITS DNA sequencing support Trypanosoma evansi and Trypanosoma equiperdum as subspecies or even strains of Trypanosoma brucei. Infection, Genetics, and Evolution 41: 56–62. doi: 10.1016/j.meegid.2016.03.022
Westenberger, S. J., C. Barnabé, D. A. Campbell, and N. R. Sturm. 2005. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171: 527–543. doi: 10.1534/genetics.104.038745
Xavier, S. C. C., A. L. R. Roque, D. Bilac, D., V. A. de Araújo, et al. 2014. Distantiae transmission of Trypanosoma cruzi: A new epidemiological feature of acute Chagas disease in Brazil. PLoS Neglected Tropical Diseases 8: e2878. doi: 10.1371/journal.pntd.0002878
Xavier, S. C. C., A. L. R. Roque, V. dos S. Lima, K. J. Monteiro, et al. 2012. Lower richness of small wild mammal species and Chagas disease risk. PLoS Neglected Tropical Diseases 6: e1647. doi: 10.1371/journal.pntd.0001647
Yaro, M., K. A. Munyard, M. J. Stear, and D. M. Groth. 2016. Combatting African Animal Trypanosomiasis (AAT) in livestock: The potential role of trypanotolerance. Veterinary Parasitology 225: 43–52. doi: 10.1016/j.vetpar.2016.05.003
Ziccardi, M., R. L. de Oliveira, M. C. Alves, and M. de F. F. da Cruz. 2005. Trypanosoma saimirii Rodhain, a junior synonym of Trypanosoma rangeli Tejera. Journal of Parasitology 91: 653–656. doi: 10.1645/GE-408R
Zingales, B., S. G. Andrade, M. R. Briones, D. A. Campbell, et al. 2009. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Memórias do Instituto Oswaldo Cruz 104: 1,051–1,054. doi: 10.1590/S0074-02762009000700021
Supplemental Reading
Cantillo-Barraza, O., S. C. Bedova, S. C. C. Xavier, S. Zuluaga, et al. 2020. Trypanosoma cruzi infection in domestic and synanthropic mammals such as potential risk of sylvatic transmission in a rural area from north of Antioquia, Colombia. Parasite Epidemiology and Control 11: e00171. doi: 10.1016/j.parepi.2020.e00171
Noun
Plural: flagellums or flagella
From Latin: flagellum = whip
Definition 1: Any of various whiplike appendages
Definition 2: A protoplasmic process, longer than a cilium, whose movements usually effect locomotion of the cell
Definition 3: The whip-like tip of the male copulatory organ in some invertebrates
Definition 4: Among crustaceans, the multiarticulate distal portion of the antennule, antenna, or exopod
Definition 5: Among insects, the distal portion of the antenna beyond the second segment (pedicel)
Definition 6: Among Porifera, a long projection from a cell, used as a propeller
Noun, plural
Singular: mitochondrion
From Greek: mitos = thread; chondros = grain
Definition: Sausage-shaped structures in the cytoplasm of animal and plant cells
Noun
Definition: Among trypanosomes, the place in the body from which the flagellum originates
Definition: A parasite that cannot exist without a host during all or some portion of the life cycle. see facultative parasite
Adjective
From Greek: monos = one; xenos = guest/host/stranger
Definition: Living within a single host during a parasite's life cycle
Adjective
From Greek: heteros = different; xenos = host/guest/stranger
Definition: Having more than one host during a parasite's life cycle
Definition: A definitive host in which the infection usually resides in nature
Noun
Plural: proboscises
From Greek: proboskis = trunk
Definition 1: Any extended trunk or beaklike sucking mouth parts of numerous invertebrates, as of leeches, planarians, dipteran insects, nemertine worms, acanthacephalans, annelids and molluscs
Definition 2: Among echinoderms, the muscular food gathering and respiratory organ extending from the trunk near the mouth
Noun
From Latin: cibarius = pertaining to food
Definition: Among insects, the part of the pre-oral cavity enclosed by the hypopharynx and the clypeus; the sucking pump in hemipterans
Noun
Adjective: monomorphic
From Greek: monos = one; morphe = form
Definition 1: A population that exhibits a single form
Definition 2: Species that contain only the female sex
Definition 3: Among social insects, having within a species or colony only a single worker subcaste
Adjective
From Greek: meta = after; kyklos = circle
Definition: Pertaining to a stage in the life cycle of a parasite that is infective to its definitive host
Adjective
Also spelled pleiomorphic
From Greek: pleion = more; morphe = form
Definition: Having the ability to change shape; polymorphic, or a type of polymorphism
Definition: The complete series of successive forms through which any particular kind of organism passes in the course of its development to maturity

