9 Epigenetics: A Healthy Life Starts Earlier Than You Think
Sabine Zempleni and Meryn Potts

Epigenetics is a field of research that started to receive increasing attention at the end of the 1990s and has taken off during the last two decades. Today, it is accepted that epigenetic changes due to environmental factors such as an unhealthy or sedentary lifestyle, obesity, excessive stress, and aging will influence our health throughout life.
This is the reason I added this chapter. You will only fully understand how nutrition and lifestyle contribute to the health of current and future generations if you understand the basics of epigenetics.
Epigenetics is a complex molecular concept, and at the moment we only understand the big ideas and a number of isolated mechanisms. Imagine the situation as a huge puzzle. We know what the picture will most likely look like, but there are many disconnected islands of puzzle pieces. It will take a lot of work by scientists to fully understand how obesity and lifestyle influence the epigenome, and more importantly, what we can do about it.
Here is an infographic reviewing the major concepts you need to understand in order to learn about epigenetics. I added the URL for some videos at the end of the chapter as well.
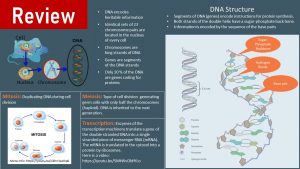
You Will Learn:
- The epigenome regulates which genes are expressed and which ones are not.
- The epigenome organizes the genome by wrapping the DNA around histones.
- Euchromatin: Packaging is loose, switching genes on.
- Heterochromatin: Packaging is tight, switching genes off.
- Packaging is regulated by adding or removing epigenetic marks. The marks can be attached directly to the DNA or to the histone tails.
- The diet can modify the epigenome by supplying:
- the epigenetic marks
- molecules that influence the epigenetic enzyme machinery
- Changes to the epigenome during the sensitive periods can be inherited by children and grandchildren.
- Your epigenome changes profoundly throughout life.
The Epigenome Regulates Gene Expression

You probably know several identical twins. Identical twin babies look so similar that parents often tie ribbons around their ankles to tell them apart. As the twin babies grow, they start looking slightly different. The older the twins get the more pronounced the differences are. This is measurable as well: 30% of identical twins end up with a different height.
Twin studies are popular in science to distinguish if a condition is due to genetics (identical for both twins) or environmental influences. Researchers studying breast cancer found something surprising. Both twins had genetic markers (the “genotype”) increasing the risk for breast cancer, but only one twin developed the disease (the “phenotype”) during their lifetime. Something did not make sense here: If the DNA is the sole blueprint for our body, then this should not happen. If both twins have a genetic disease marker, both twins should develop the disease.
This was not the only riddle. Humans start out with a single cell, the zygote, that starts dividing. The zygote is a totipotent cell that can develop into any cell type. All daughter cells of the zygote have a copy of the genome organized in 23 pairs of chromosomes. The chromosomes are built from 3.2 billion base pairs. Interestingly, only 1-2% of the base pairs code for an estimated 50,000 genes. Of those 50,000 genes, only one tenth is turned on in any given cell type.
Something tells one cell to function as a brain cell, another as a skin cell, another as a muscle cell, bone cell, liver cell and so forth. The DNA in each cell type is identical but this “something” silences 90% of the genes that are not needed for the function of the cell.
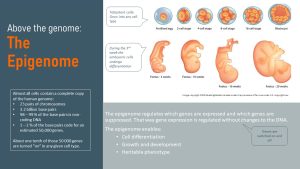
This “something” is the epigenome. Epi is Greek and means above, in this case above the genome. The epigenome regulates which genes are expressed and which ones are not without changing the genetic code.
Turning sections of the genome off and allowing specific genes to stay on allows a stem cell, the totipotent cell, to become and function as a brain cell. Epigenetic switches also explain the phenotype of a person, the observable characteristics such as skin and hair color, size, and biochemical properties. Just like the genome, most of the epigenome is heritable.
The Epigenome Organizes the Genome by Wrapping the DNA Around Histones
If you want to understand how those epigenetic switches work, you need to understand how DNA is organized. We tend to think of the genes as Xs, but genes appear in this form only during mitosis.
DNA is also not a long, stringy double helix bunched up in the nucleus; it is packaged in an elaborate way, allowing for six feet of DNA to fit into six µm of nucleus.
Below is a video (watch until 1:17) explaining DNA packaging. You can also access the video through copy/pasting the following link: https://youtu.be/OjPcT1uUZiE.
DNA is wrapped around proteins called histones. Stretches of DNA can be wrapped loosely or tightly, and this determines if genes are “on” or “off.”
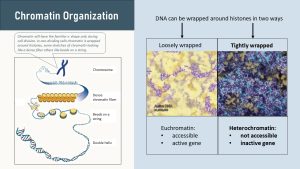
If the stretch of DNA is loosely wrapped around the histones, the gene is switched on. Enzymes can easily unravel the DNA from the histone allowing transcription. Enzymes read the piece of unravelled DNA producing a strand of messenger RNA. Loosely wrapped, accessible genes are called euchromatin. Euchromatin has the “beads on a string” appearance seen above (loose purple beads on a yellow background).
If genes are wrapped tightly (purple side in above picture), genes are not accessible to the transcription enzymes. The gene is “turned off.” This tight wrapping of DNA around the histones is called heterochromatin.
How can DNA be wrapped tightly or loosely? Watch the following video starting at 5:10 min.
Histones are negatively charged proteins that are attracted to the positively charged DNA. The degree to which DNA is tightly or loosely bound to histones is modified by the addition of epigenetic marks to the DNA or the tails of the histones. Epigenetic marks are small molecules interacting with neighboring molecules. Depending on the type of epigenetic mark, the bond between histone and DNA will weaken or strengthen.
Epigenetic Marks Determine the Packaging Of the DNA and Can Be Attached Directly to the DNA or the Histone Tails
There are two types of epigenetic marks. Adding a methyl group directly to the DNA or attaching small functional groups to histone tails.
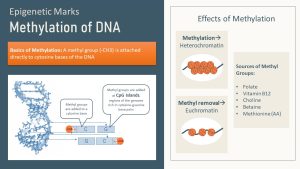
The first type of epigenetic marks are methyl groups attached to the DNA. Methylation of DNA is the most advanced research area when it comes to the epigenome. The methyl groups are mostly attached to specific regions of the DNA known as “CpG islands.” CpG islands are sections of the DNA rich in cytosine (C) and guanine (G) base pairs. The methyl group is added enzymatically to the cytosine.
The second type of epigenetic mark is the attachment of a small functional molecule to a histone tail. Envision the histone proteins as having amino acid chains protruding from the nucleosome (see the picture below as an example). Enzymes can attach a variety of functional groups to those tails. The most common molecules serving as epigenetic marks on histone tails are acetyl, methyl, or phosphor groups.
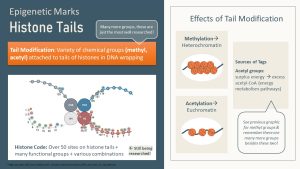
Many chemical groups can be used as epigenetic marks though, and there are over 50 different sites for epigenetic mark attachment on the histone tail. Not to mention that enzymes can attach just one mark or several at the same time. You can imagine the huge amount of epigenetic mark combinations that can occur. Today, only a few select sites or combinations are researched, and much of the histone code is not identified.
Despite this overall uncertainty, we do know some mechanisms:
DNA and histone tail methylation will often compact a gene, turning that stretch of DNA into heterochromatin (switching off).
On the other hand, acetylation of histone tails tends to loosen heterochromatin and the gene becomes switched on. When acetyl groups are attached the positive charge of the histone is reduced or removed. A reduced positive charge decreases the attraction between histones and the negatively charged DNA.
Overall, the compacting or unlocking of genes is much more complicated, involving DNA methylation, histone tail modifications, and various other proteins. Again, the exact mechanisms are not entirely known yet.
Non-coding RNAs are also considered part of the epigenome. They are not epigenetic marks but they modify DNA after transcription has taken place.
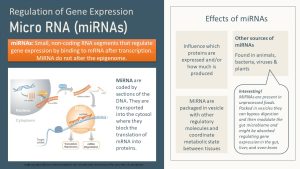
Especially interesting for us is the group of the micro-RNA or miRNA for short(er). MiRNAs do not alter the epigenetic state, but they suppress or fine tune gene expression after transcription has taken place.
Short pieces of RNA are transcribed, transported out of the nucleus into the cytosol, and enzymatically shortened. Those short pieces of RNA then bind to a matching mRNA segment, blocking the translation of the mRNA into protein(s). MiRNA therefore influence which proteins are expressed and how much is produced.
Even more interesting, miRNA are packaged into vesicles and secreted into the bloodstream to orchestrate gene expression in other tissues. MiRNA is the texting system of the body. Muscle talking to the liver, adipose tissue talking to brain, liver, pancreas, and muscles.
New research shows that miRNAs packaged in vesicles can be found in unprocessed foods. These vesicles can bypass digestion, and the vesicle cargo (for example, miRNA) can modulate the gut microbiome. Scientists have found these food vesicles in the liver and brain. While this exciting research is brand new, it indicates that the food we eat may modulate our metabolism and health.
Let’s summarize before looking into how eating choices might affect the epigenome:
1. Genes are suppressed or expressed by winding the corresponding piece of DNA around histones.
- Euchromatin: DNA wound loosely; gene accessible to enzymes and “switched on”
- Heterochromatin: DNA wound tightly; gene inaccessible to enzymes and “switched off”
2. Modulating how tight chromatin is packaged depends on the combination of epigenetic marks attached:
- Methyl groups attached to the DNA (tend to package tightly and turn genes off)
- Histone tail modification: Variety of chemical groups attached in various combinations to histone tails
3. The epigenome is regulated by:
- Epigenetic machinery (enzymes)
- Availability of epigenetic marks
- Non-coding RNA (post-translational)
The Diet Supplies Epigenetic Marks and Regulates the Epigenetic Enzyme Machinery
To understand how our diet modulates the epigenome, you need to separate the two components of the epigenetic regulation:
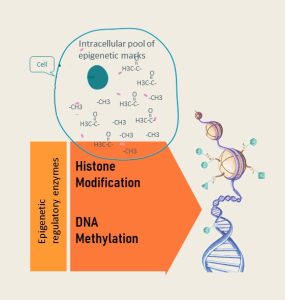
- Pool of epigenetic marks (sometimes called tags) in the cell: Recall that the expression of the DNA is regulated by how tightly the double helix is wound around histones. This is accomplished by attaching epigenetic tags either directly to the DNA (DNA methylation) or attaching epigenetic tags to the histone tails (histone modification). Every time a cell divides the tags need to be duplicated as well. If for example the methyl group pool in the cell is low, DNA methylation cannot be replicated in the new daughter cell. The sections with low DNA methylation might switch on unwanted genes.
- Epigenetic machinery: Epigenetic tags are attached by a set of enzymes. The amount and activity of enzymes can vary. Their expression and activity can be up- or down-regulated by signaling molecules and is fine-tuned by micro-RNA. Collectively, this is called the epigenetic machinery.
So far, 1) DNA methylation, and 2) histone tail methylation/acetylation are the most researched epigenetic modifications. From this research, we know that DNA methylation tends to turn sections of DNA into heterochromatin, turning a gene off. Acetyl groups attached to the histone tails tend to turn genes on.
The diet impacts the epigenome in three ways:
- Ensuring that the intracellular methyl group pool is adequate
- Supplying a variety of other epigenetic marks (tags)
- Modulating the enzymes attaching the marks
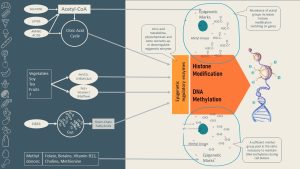
Let us start with the pool of molecules that can be used as epigenetic marks:
Methyl groups come from two vitamins that serve as dietary methyl donors: folate and vitamin B12. (For example, research shows that newborns of women with folic acid supplementation during pregnancy have higher levels of DNA methylation!) Other methyl donors include the amino acid methionine, as well as betaine and choline.
*For specific foods that contain these nutrients, refer to the graphic in the following section: Is a Choline-Rich Diet the Key to Everlasting Life?*
The second group of marks are acetyl groups, which come from the energy metabolism. All surplus glucose and amino acids (plus alcohol) from the diet are fed into metabolic pathways in the liver until they are metabolized to acetyl-CoA. Acetyl-CoA is then used for lipogenesis. This means that over-nutrition tends to produce an acetyl-CoA surplus in the cell. It is hypothesized that this acetyl-CoA surplus promotes an acetylation of histone tails and turns genes on. Starvation and excessive exercising, however, will reduce the availability of acetyl marks. Lipids are different: surplus fat is directly stored as fat in the adipose tissue, but lipolysis produces lots of acetyl-CoA.
In addition to epigenetic mark availability, diet can also influence the activity of the epigenetic enzyme machinery (remember that enzyme activity can be up- or down-regulated). This area is much less understood. Scientists think that metabolites from the citric acid cycle, phytochemicals, and some nutrients regulate the epigenetic machinery. The most researched phytochemicals are the ones from tea, cruciferous plants, and soy, as well as resveratrol from blue and red fruits and vegetables. The field is rapidly expanding and will be exciting to watch.
The newest research area looking into the relationship between diet and epigenetic regulation of the genome involves the gut microbiome. New research suggests that diet composition modulates the gut microbiome, and the resulting metabolites produced by the microbes then modulate the epigenetic machinery. For example, if the diet supplies enough fiber, the microbiota will produce ample amounts of short-chain fatty acids. Those SCFA modulate the epigenetic enzyme machinery. Scientists hypothesize that this is one explanation of how diet and cancer are connected.
Just looking at the infographic above and thinking about the many ways the diet can vary between individuals, it should become clear how complicated it is to research the connection between diet and epigenetic regulation of DNA expression. This level of complexity will require innovative research approaches and collaborations between different branches of science. Nutritionists and biochemists may have the right knowledge, but will not have the tools to research this level of complexity. It will require involvement of data scientists, programmers, and biological systems engineers—just to name a few—to make progress.
Is a Choline-Rich Diet the Key to Everlasting Life? Not so Fast, it is More Complicated Than That
Epigenetics have appeared in the media and influencer spheres in the last several years. The main hypothesis journalists and influencers latched onto is that a diet rich in choline (a major methyl donor) will promote methylation of DNA. Well-methylated DNA is supposed to suppress genes that code for genetic predisposition of chronic diseases such as T2D, CVD, and certain cancers. These authors then suggest that eating a diet rich in animal products provides a lot of choline and is therefore beneficial for health. How much of this is evidence-based?
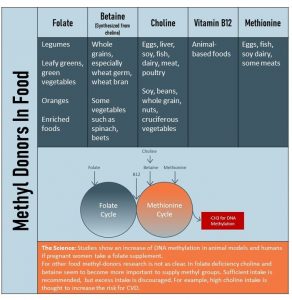
The table above lists methyl-donors and their food sources. A mixed, healthy diet will provide plenty of methyl donors. The problem is that folate and choline intake, both essential nutrients, is on the lower side in the US. The goal for everybody is a sufficient dietary intake of folate, B12, and choline from a healthy diet.
Newer studies caution that too much choline, predominantly from animal food sources, is connected to an increased risk for CVD. Choline is metabolized by gut bacteria and the metabolite is converted to trimethylamine N-oxide (TMAO). We do not know if high TMAO blood concentrations contribute to atherosclerosis and heart disease, but people with high TMAO concentrations do have a higher risk.
 Now to the evidence: Most of the research in this area was completed during pregnancy. A landmark study showed that mice fed a diet with added choline during pregnancy resulted in pups that were less likely to be obese as adult mice.
Now to the evidence: Most of the research in this area was completed during pregnancy. A landmark study showed that mice fed a diet with added choline during pregnancy resulted in pups that were less likely to be obese as adult mice.
This sounds great, but it is not that easy. The mice used in this study are called Agouti mice (picture to the left [1]) Agouti mice have a specific gene that makes them prone to obesity. The same gene also codes for their coat color.
If the Agouti gene is switched on the mouse is yellow and becomes obese very rapidly. If the gene is switched off the mouse is brown and lean. Adding choline to the mother’s diet switches the gene off and results in thin brown mice offspring. This effect lasted through two generations of mice.
Obesity epidemic solved? Not even close. Keep in mind that specific Agouti mice were used. The study demonstrated elegantly that genes can be switched on and off by feeding a methyl-rich diet and that those changes were inherited to the next generations. The study cannot prove that a methyl-rich diet will prevent obesity in humans.
After this landmark study, many scientists started researching this hypothesis in animals as well as pregnant women and confirmed that a sufficient supply of methyl groups during pregnancy results in a higher degree of methylated DNA in the fetus. The most advanced research in humans shows that folate supplementation is connected with higher DNA methylation in babies. The connection to obesity remains elusive so far.
Other landmark observational studies looked at the connection between starvation or times of abundance and health outcomes in the next two generations. The results were much more inconsistent but indicated that starvation and over-nutrition have a negative impact on health for the next generations.
Today, research takes place in many areas piecing together our understanding of the epigenome, factors influencing the epigenome, and the connection to health.
The current evidence supports the following big ideas:
- Supplying sufficient methyl groups during pregnancy—many American women have a diet low in methyl-donors—might be connected to better health for the following generations. These methyl groups can come from choline-rich animal products, but studies also showed higher DNA methylation when folate was supplemented.
- Phytochemicals from a plant-based diet and sufficient essential nutrients seem to be beneficial in maintaining health. This effect might reach into the future for the next generations.
- For most of those big ideas we are only at the beginning of understanding the underlying mechanisms.
- Over-nutrition and starvation both increase the risk for chronic diseases in following generations. The evidence is more inconsistent.
What does this mean for eating decisions? The current evidence supports the recommendation to eat a healthy, mixed, plant-based diet and maintain a healthy weight. Current evidence does not support a high meat (choline-rich) diet or even less genotype or phenotype specific dietary pattern as sometimes claimed in the media.
Changes to the Epigenome During the Sensitive Periods Can Be Inherited to Children and Grandchildren
You know now that our diet has the potential to change epigenetic marks and influence the epigenetic enzyme machinery. This, in turn, can change which genes are switched on and which genes are switched off. Lastly, there are studies that connect epigenetic changes to the risk for chronic diseases.
It does not stop there! Recall that hunger studies indicate starvation might influence metabolic health of children and even grandchildren. How does this happen?
Every time a cell divides, most of the epigenetic marks are removed; however, after cell division is completed, those marks are re-established in the daughter cell in the same pattern as before. We do not yet know how those marks are mitotically inherited from cell to cell, only that they are. We also know that over a lifetime epigenetic marks change. For example, methylation of DNA declines in a pattern that scientists would like to use to determine the metabolic age of a person.
It is also thought that the epigenome reacts to our environment to adapt our body to the challenges the world throws at us. Most of those changes are not inheritable. Wait! Didn’t we just say that lifestyle decisions, especially over- and under-nutrition, can change chronic disease risk for children and grandchildren?
There are three exceptions. To understand how our lifestyle choices can alter the epigenome of our children and grandchildren, we need to look at the epigenome during the early embryonic development. There are sensitive periods when the embryo is vulnerable.
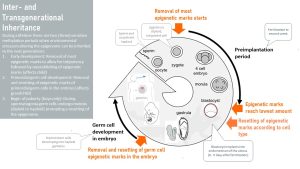
The first sensitive period occurs when the sperm meets an oocyte, fertilization takes place, and a zygote forms and starts dividing. Those cells need to develop into all sorts of tissues and the cells need to be totipotent; this is achieved by deleting most of the epigenome.
After the blastocyst implants into the lining of the placenta different cells are assigned to different tissues. In the process, the epigenetic marks are reset to establish an epigenome for different tissue types as inherited by the parents.
This time of deleting and re-establishing the epigenome according to the blueprint is called a sensitive period. External influences during this short window in the life of an embryo such as over- or undernutrition of the mother or a diet low in methyl donors, can change the resetting of the epigenome. Once the epigenome is established it will be duplicated during each cell division. On a side note, random failure to establish correct epigenetic marks will lead to the loss of the embryo, some childhood cancers, or epigenetic diseases.
The second sensitive period also takes place in-utero. This time, future grandchildren are affected.
During embryonic and fetal development, the reproductive system is created. As part of this development, cells grow into primordial germ cells that will later develop into oocytes and sperm cells. Cells destined to be germ cells need to go from a diploid cell to a haploid cell. During this process most of the epigenetic tags are removed and then re-established in the germ cell. This second sensitive period takes place during mid-gestation and is also sensitive to epigenetic mark changes. The lifestyle of the pregnant mother will now influence the metabolic health of a potential future grandchild.
The third sensitive period happens only in boys. Girls are born with developed oocytes and undergo the entire germ cell development in utero. In boys the germ cell goes dormant until the boy starts producing sperm during puberty. Early puberty in boys is the last time when epigenetic marks are mostly removed. The boy’s lifestyle—availability of methyl donors, over-or undernutrition— might influence the resetting of epigenetic marks and therefore the health of his future child.
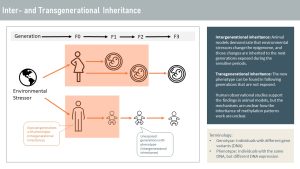
Inheritance of altered methylation patterns is mostly researched in animal models. Those models include fruit flies and worms, as well as rats and mice. During the sensitive periods, the embryo and developing germ cells are exposed, and the methylation pattern is altered. Those animal studies also demonstrated that the altered methylation pattern, and therefore the phenotype, can be inherited by the next generation that was never exposed. Today, scientists are puzzled how the altered epigenome can be inherited since the epigenome is almost completely deleted and re-established during the sensitive periods.
Animal studies are supported by studies following several generations of humans. Environmental stressors such as severe undernutrition, overnutrition, high intake of sugar, stress, environmental toxins, and more impact the metabolism and health of the following generations. It is thought that those epigenetic alterations are meant to adapt the next generation to a new environment. For example, hunger studies show that the alteration in energy metabolism allows the next generation of lab animals to survive better during famine, but those alterations predispose the animals to obesity and chronic diseases.
The Epigenome Changes Profoundly Throughout Life
Remember that I previously mentioned during cell division the epigenome is duplicated so the daughter and mother cell are identical. This is generally true, but not completely.
Based on the evidence we have today, we can hypothesize that the epigenome responds to our lifestyle and the environment we are living in. This seems to be the purpose of the epigenome.
From studies looking at the epigenome throughout life in lab animals, we know that the epigenome changes profoundly. We also see changes in humans, and this explains why identical twins start out fairly identical but then start becoming their own epigenetic person (phenotype), resulting in a slightly different look and different disease risk, for example.
These continuous changes—starting already in-utero—are not inheritable to the next generation.
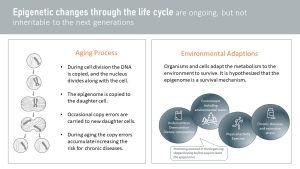
Epigenetic changes throughout life can happen two ways:
- Part of the aging process: No biological process is perfect. During cell division, random errors are made while copying epigenetic marks from the mother to the daughter cell. If the error is grave, apoptosis (cell death) is induced, but smaller mistakes will be carried forward when the daughter cell starts dividing. For example, an error during epigenetic mark duplication might switch on the gene coding for a cancer predisposition. These errors accumulate with age, and it is thought that this is one of the reasons for an increased disease risk during aging. Research in this area hopes to reset the epigenome and extend quality life (more in the The Aging Process Explained)
- Environmental adaptations: Organisms need to adapt to their environment if they want to survive in changing conditions. For example, living in an environment where infections and parasites are common, the energy metabolism needs to adapt to provide the immune system with plenty of energy. Switching a gene on that increases insulin resistance will allow the metabolism to direct glucose to the immune system. During undernutrition, insulin resistance will also help divert glucose from insulin dependent tissues to important organs such as the brain. Other research areas are physical activity, chronic diseases, excessive and chronic stress, and environmental factors such as cold exposure. Recently, research found surprising results connecting some environmental chemicals (such as DDT, BPA, glyphosates and many more) to a methylation pattern that increases the risk for obesity.
Epigenome research is exciting, and will enable us to explain many hypotheses regarding chronic disease that we have not been able to explain so far; however, we should keep in mind that (at this point) the evidence resembles a big puzzle with many gaps between islands of connected pieces:

Interested In More Information?
- Mitosis: https://youtu.be/C6hn3sA0ip0: https://youtu.be/C6hn3sA0ip0
- Transcription: https://youtu.be/SMtWvDbfHLo
- Fessele KL, Wright F. Primer in Genetics and Genomics, Article 6: Basics of Epigenetic Control. Biol Res Nurs. 2018;20(1):103-110. doi:10.1177/1099800417742967
Editors: Meryn Potts (infographics and editing), Eric Hanzel, Gabi Ziegler
NUTR251 Contributors:
- Spring 2020: Kennedie Engles, Caleb Licking, Alexander Casebeer, Elliot Glaser, Hailie Slepicka, Rose Davidson, Bryce Heiser, Allison Aden, Kyle Dawson, Grace Neville, Madison Yourstone, Bryce Heiser, Melanie Nissen, Cameron Hucke
- Image copyright 2011 https://www.sciencedirect.com/topics/neuroscience/agouti-gene. Included under the provisions of fair use under U.S. copyright law. ↵
Type of cell division that results in two daughter cells with the same chromosomes as the parent cell, typical during ordinary tissue growth.
Chromatin: The complex of DNA and proteins
A modified amino acid: Glycin plus three methyl group. The attached methyl groups make betaine a methyl donor
Two sets of chromosomes
Single set of chromosomes


Feedback/Errata